Running a K-means Cluster Analysis
 Kamya Kumari
Kamya KumariWhat is K-means Clustering
K-means clustering is the unsupervised machine learning algorithm that is part of a much deep pool of data techniques and operations in the realm of Data Science. It is the fastest and most efficient algorithm to categorize data points into groups even when very little information is available about data.
More on, similar to other unsupervised learning, it is necessary to understand the data before adopting which technique fits well on a given dataset to solve problems. Considering the correct algorithm, in return, can save time and effort and assist in obtaining more accurate results.
Explanation: Working of K-means Algorithm
By specifying the value of k, if k is equal to 3, the algorithm accounts for 3 clusters. Following are the working steps of the K-means Algorithm :
- K-center are modeled randomly in accordance with the present value of K.
- K-means assigns each data point in the dataset to the adjacent center and attempts to curtail Euclidean distance between data points. Data points are assumed to be present in the peculiar cluster as if it is nearby to center to that cluster than any other cluster center.
- After that, k-means determines the center by accounting for the mean of all data points referred to that cluster center. It reduces the complete variance of the intra-clusters with respect to the prior step. Here, the “means” defines the average of data points and identifies a new center in the method of k-means clustering.
- The algorithm gets repeated among steps 2 and 3 till some paradigm will be achieved such as the sum of distances between data points and their respective centers are diminished, an appropriate number of iterations is attained, no variation in the value of cluster center or no change in the cluster due to data points.
Example Code are given below to get a response on a set of clustering variables:
from pandas import Series, DataFrame
import pandas as pd
import numpy as np
import matplotlib.pylab as plt
from sklearn.model_selection import train_test_split
from sklearn import preprocessing
from sklearn.cluster import KMeans
from scipy.spatial.distance import cdist
from sklearn.decomposition import PCA
import statsmodels.formula.api as smf
import statsmodels.stats.multicomp as multi
%matplotlib inline
RND_STATE = 55121
1. Loading Data Code
data = pd.read_csv("data/tree_addhealth.csv")
data.columns = map(str.upper, data.columns)
1.1 Removing rows with incomplete data
data_clean = data.dropna()
1.2 Selecting clustering variables
cluster=data_clean[['ALCEVR1','MAREVER1','ALCPROBS1','DEVIANT1','VIOL1',
'DEP1','ESTEEM1','SCHCONN1','PARACTV', 'PARPRES','FAMCONCT']]
cluster.describe()
2. Preprocessing Data
2.1 Scaling Data
clustervar=cluster.copy()
clustervar['ALCEVR1']=preprocessing.scale(clustervar['ALCEVR1'].astype('float64'))
clustervar['ALCPROBS1']=preprocessing.scale(clustervar['ALCPROBS1'].astype('float64'))
clustervar['MAREVER1']=preprocessing.scale(clustervar['MAREVER1'].astype('float64'))
clustervar['DEP1']=preprocessing.scale(clustervar['DEP1'].astype('float64'))
clustervar['ESTEEM1']=preprocessing.scale(clustervar['ESTEEM1'].astype('float64'))
clustervar['VIOL1']=preprocessing.scale(clustervar['VIOL1'].astype('float64'))
clustervar['DEVIANT1']=preprocessing.scale(clustervar['DEVIANT1'].astype('float64'))
clustervar['FAMCONCT']=preprocessing.scale(clustervar['FAMCONCT'].astype('float64'))
clustervar['SCHCONN1']=preprocessing.scale(clustervar['SCHCONN1'].astype('float64'))
clustervar['PARACTV']=preprocessing.scale(clustervar['PARACTV'].astype('float64'))
clustervar['PARPRES']=preprocessing.scale(clustervar['PARPRES'].astype('float64'))
2.2 Splitting into train test splits
clus_train, clus_test = train_test_split(clustervar, test_size=0.3, random_state=RND_STATE)
3. Making K-means analysis for 1 to 9 clusters
clusters=range(1,10)
meandist=[]
for k in clusters:
model=KMeans(n_clusters=k)
model.fit(clus_train)
clusassign=model.predict(clus_train)
meandist.append(sum(np.min(cdist(clus_train, model.cluster_centers_, 'euclidean'), axis=1))
/ clus_train.shape[0])
4. Plotting relation between the number of clusters and average distance
plt.plot(clusters, meandist)
plt.xlabel('Number of clusters')
plt.ylabel('Average distance')
plt.title('Selecting k with the Elbow Method')
plt.show()
Output of Plot:
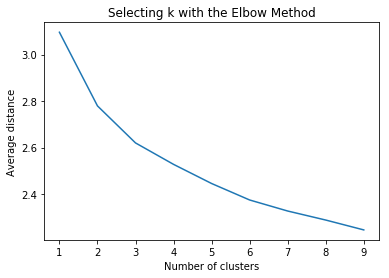
5. Checking solution for 3 clusters model
model3=KMeans(n_clusters=3)
model3.fit(clus_train)
clusassign=model3.predict(clus_train)
5.1 Plotting clusters
pca_2 = PCA(2)
plot_columns = pca_2.fit_transform(clus_train)
plt.scatter(x=plot_columns[:,0], y=plot_columns[:,1], c=model3.labels_,)
plt.xlabel('Canonical variable 1')
plt.ylabel('Canonical variable 2')
plt.title('Scatterplot of Canonical Variables for 3 Clusters')
plt.show()
Output for Plotting Cluster
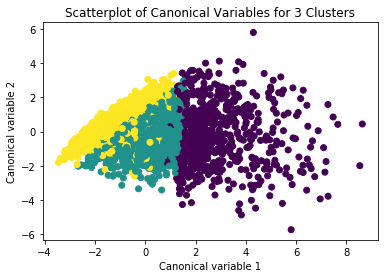
5.2 Merging cluster assignment with clustering variables to examine cluster variable means by cluster
clus_train.reset_index(level=0, inplace=True)
cluslist=list(clus_train['index'])
labels=list(model3.labels_)
newlist=dict(zip(cluslist, labels))
newclus=DataFrame.from_dict(newlist, orient='index')
newclus.columns = ['cluster']
newclus.describe()
| cluster | |
| count | 3202.000000 |
| mean | 1.232042 |
| std | 0.768115 |
| min | 0.000000 |
| 25% | 1.000000 |
| 50% | 1.000000 |
| 75% | 2.000000 |
| max | 2.000000 |
newclus.reset_index(level=0, inplace=True)
merged_train=pd.merge(clus_train, newclus, on='index')
merged_train.head(n=100)
merged_train.cluster.value_counts()
5.3 Calculating clustering variable means by cluster
clustergrp = merged_train.groupby('cluster').mean()
print ("Clustering variable means by cluster")
print(clustergrp)
5.4 Validating clusters in training data by examining cluster differences in GPA using ANOVA
gpa_data=data_clean['GPA1']
gpa_train, gpa_test = train_test_split(gpa_data, test_size=.3, random_state=RND_STATE)
gpa_train1=pd.DataFrame(gpa_train)
gpa_train1.reset_index(level=0, inplace=True)
merged_train_all=pd.merge(gpa_train1, merged_train, on='index')
sub1 = merged_train_all[['GPA1', 'cluster']].dropna()
gpamod = smf.ols(formula='GPA1 ~ C(cluster)', data=sub1).fit()
print (gpamod.summary())
print ('means for GPA by cluster')
m1= sub1.groupby('cluster').mean()
print (m1)
print ('standard deviations for GPA by cluster')
m2= sub1.groupby('cluster').std()
print (m2)
mc1 = multi.MultiComparison(sub1['GPA1'], sub1['cluster'])
res1 = mc1.tukeyhsd()
print(res1.summary())
Subscribe to my newsletter
Read articles from Kamya Kumari directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
