Exploring Stereochemistry with RDKit's EnumerateStereoisomers.py
 Biohacker0
Biohacker0Table of contents

In the last blog, we discussed the principle, now we will go through the GitHub code of it.
Introduction:
I read the whole workings of EnumerateStereoisomers function, what’s the mechanism, and what rules are followed while making these stereoisomers and went through the whole code line by line on GitHub and understood what each function and class did
Abstract Working's:
so we have some classes and methods that are doing their tasks and pass the processed values to others, let's see what each of them does:
StereoEnumerationOptions Class:
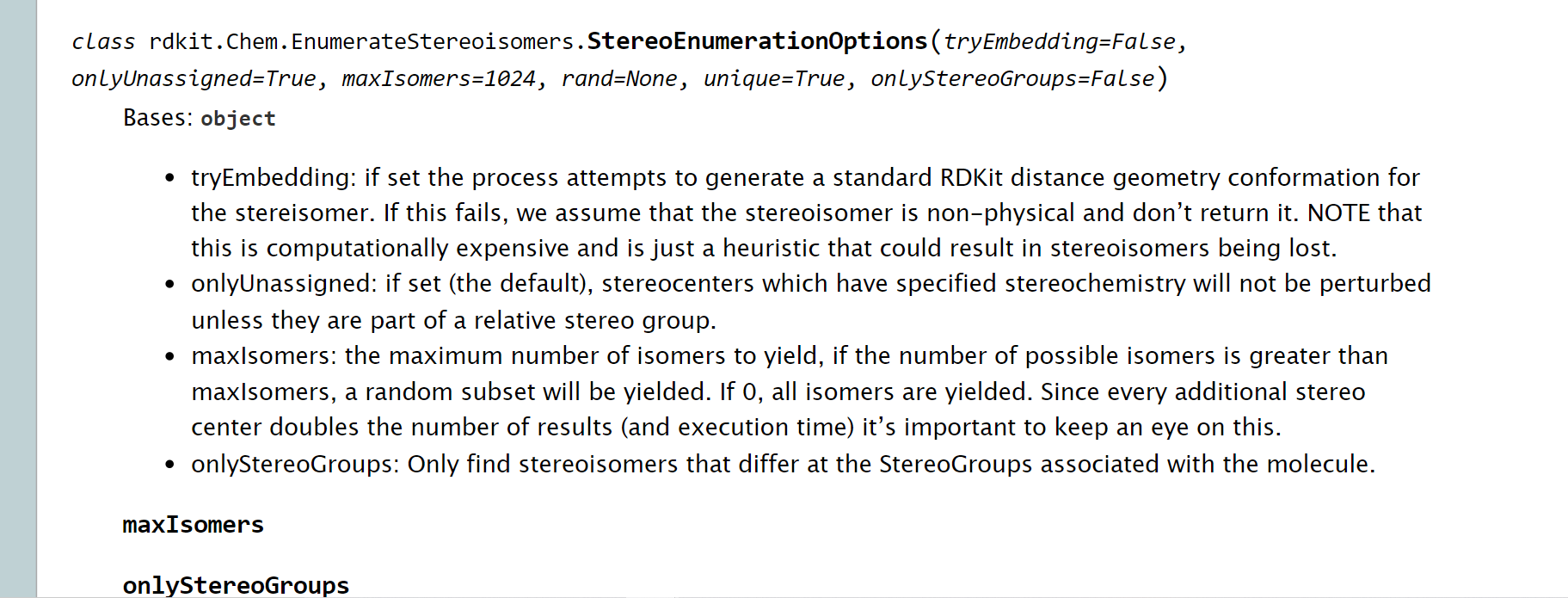
Purpose: This class defines various options and parameters for stereoenumeration.
Parameters:
tryEmbedding: If set, it attempts to generate a 3D conformation for each stereoisomer.onlyUnassigned: Controls whether only unspecified stereocenters are perturbed.onlyStereoGroups: Restricts the enumeration to stereoisomers that differ in stereo groups.maxIsomers: Sets the maximum number of isomers to be generated.rand: An optional random number generator for controlling isomer sampling.unique: If set, ensures that only unique isomers are generated.
_BondFlipper, _AtomFlipper, _StereoGroupFlipper Classes:
Purpose: These classes are used to flip the stereochemistry of bonds, atoms, and stereo groups, respectively.
_BondFlipper.flip,_AtomFlipper.flip,_StereoGroupFlipper.flip: Methods to change the stereo configuration of the associated element.
_getFlippers Function:
Purpose: Determines which elements (bonds, atoms, and stereo groups) can have their stereochemistry flipped.
Identifies and collects elements that can change stereochemistry based on user-defined options.
_RangeBitsGenerator, _UniqueRandomBitsGenerator Classes:
Purpose: These classes generate bit patterns to represent different stereoisomer configurations.
_RangeBitsGenerator: Generates all possible bit patterns._UniqueRandomBitsGenerator: Generates random, unique bit patterns within the specified limits.
GetStereoisomerCount Function:
Purpose: Provides an estimate of the total number of possible stereoisomers for a molecule.
Uses the number of potential stereo flippers (flippable stereocenters and bonds) to calculate the count.
EnumerateStereoisomers Function:
Purpose: Generates stereoisomers for a given molecule.
Uses the options and random bit patterns to enumerate different stereoisomer configurations.
Checks for uniqueness and attempts to embed 3D conformations if specified.
Yields the generated stereoisomers one by one.
Detailed Working:
EnumerateStereoisomers.py code from RDKit:
StereoEnumerationOptions Class:
This class defines various options for stereoenumeration.
tryEmbedding: If set toTrue, the code attempts to generate a 3D conformation for each stereoisomer. If embedding fails, the stereoisomer is not returned. This option can be computationally expensive.onlyUnassigned: By default set toTrue, it specifies that stereocenters with already specified stereochemistry will not be perturbed unless they are part of a relative stereo group.onlyStereoGroups: If set toTrue, the code only finds stereoisomers that differ at the stereo groups associated with the molecule.maxIsomers: Specifies the maximum number of isomers to yield. If the number of possible isomers exceeds this limit, a random subset is yielded. If set to 0, all isomers are yielded.rand: An optional parameter that allows you to provide a random number generator for controlling isomer sampling.unique: If set toTrue, ensures that only unique isomers are generated.
_BondFlipper, _AtomFlipper, _StereoGroupFlipper Classes:
These classes are used to flip the stereochemistry of different elements:
_BondFlipper: Flips the stereochemistry of a bond._AtomFlipper: Flips the stereochemistry of an atom._StereoGroupFlipper: Flips the stereochemistry of a stereo group.
Each class has a
flipmethod that changes the stereo configuration of the associated element.
_getFlippers Function:
This function determines which elements (bonds, atoms, and stereo groups) can have their stereochemistry flipped based on the input molecule and user-defined options.
It first calls
Chem.FindPotentialStereoBonds(mol)to identify potential stereocenters and bonds.It collects the elements that can change stereochemistry based on the
optionsprovided:For atoms, it collects those with unspecified or unassigned chiral tags.
For bonds, it collects those with non-STEREONONE stereochemistry, if not restricted to unassigned bonds.
For stereo groups, it collects elements that are not of type STEREO_ABSOLUTE, if not restricted to unassigned groups.
Returns a list of flippers (instances of
_BondFlipper,_AtomFlipper, or_StereoGroupFlipper) for elements that can be flipped.
_RangeBitsGenerator, _UniqueRandomBitsGenerator Classes:
These classes are responsible for generating bit patterns to represent different stereoisomer configurations.
_RangeBitsGenerator: Generates all possible bit patterns for stereoisomer configurations, ranging from 0 to 2^nCenters (where nCenters is the number of flippable elements)._UniqueRandomBitsGenerator: Generates random, unique bit patterns within the specified limits (maxIsomers).
GetStereoisomerCount Function:
This function provides an estimate of the total number of possible stereoisomers for a given molecule.
It takes the input molecule
mand theoptionsas parameters.First, it creates a copy of the molecule (
tm) and clears certain properties and bond directions.It then calls
_getFlippersto determine the number of flippable elements (nCenters).The function returns 2^nCenters as an estimate of the number of possible stereoisomers.
EnumerateStereoisomers Function:
This function is the core of stereoisomer enumeration.
It takes the input molecule
m, theoptions, and an optionalverboseflag as parameters.The function starts by clearing certain properties and bond directions of the input molecule.
It then determines the number of flippable elements (nCenters) by calling
_getFlippers.If there are no flippable elements, the input molecule itself is yielded as an isomer.
If the number of possible isomers is within the specified limit (maxIsomers), it uses
_RangeBitsGeneratorto iterate through all possible bit patterns and flip the stereochemistry accordingly.For each bit pattern, it flips the stereochemistry of elements, removes stereogroups (if present), assigns stereochemistry, and optionally embeds 3D conformations.
If the unique option is set, it ensures that only unique isomers are yielded.
The generated isomers are yielded one by one.
The function can also handle cases where embedding fails for certain isomers (controlled by the
tryEmbeddingoption).The
verboseflag controls the verbosity of output when embedding fails.
thread: link
Summary
In theory, this code enables the enumeration of stereoisomers by systematically flipping the stereochemistry of bonds, atoms, and stereo groups in the input molecule. It considers user-defined options for controlling the enumeration process, ensuring uniqueness, and attempting to embed 3D conformations. The result is a generator that yields different stereoisomer configurations of the input molecule
Subscribe to my newsletter
Read articles from Biohacker0 directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by

Biohacker0
Biohacker0
Making cool stuff