Using Conda without having to install locally
 k
k
Conda is a powerful environment manager but, much like its namesake, it can be large, slow, and unpleasant to have around. You can use rx to install and run conda in the cloud, keeping your system unbloated (while still allowing you to develop locally).
This post takes you through setting up a project that runs conda on a remote host. The whole process should take about 10 minutes (most of the time is conda installing dependencies). Feel free to use an existing project or create a new directory for this.
To get started, create a directory for your project and add an environment.yaml file (this is how conda determine which dependencies a project is using). Let's use numpy, a famously annoying install. environment.yaml should contain:
name: hello-conda
channels:
- defaults
dependencies:
- matplotlib
- numpy
Now start up the remote machine and copy your project to it with:
$ pip install run-rx # Install rx itself
$ rx init # Start a remote workspace
This will take a few minutes to set up the workspace. Because you have an environment.yaml file, rx knows that your project uses conda and sets up your machine appropriately.
Once it's done, you can try running anything you want on the remote machine by prefixing your command with "rx". For example, you can poke around and look at the filesystem:
$ rx pwd # check what directory you're in
/root/rx/app
$ rx ls # check what files were copied here
environment.yaml
Now we want to try a simple program that uses numpy. Let's create a script that plots the Mandelbrot set, mandelbrot.py. Put it in the same directory as your environment.yaml file.
# Slightly modified version of
# https://numpy.org/doc/stable/user/quickstart.html#indexing-with-boolean-arrays
import numpy as np
import matplotlib.pyplot as plt
width = 400
height = 400
def mandelbrot(w, h, maxit=20, r=2):
"""Returns an image of the Mandelbrot fractal of size (h,w)."""
x = np.linspace(-2.5, 1.5, 4*w+1)
y = np.linspace(-1.5, 1.5, 3*h+1)
A, B = np.meshgrid(x, y)
C = A + B*1j
z = np.zeros_like(C)
divtime = maxit + np.zeros(z.shape, dtype=int)
for i in range(maxit):
z = z**2 + C
diverge = abs(z) > r # who is diverging
div_now = diverge & (divtime == maxit) # who is diverging now
divtime[div_now] = i # note when
z[diverge] = r # avoid diverging too much
return divtime
result = mandelbrot(width, height)
output_file = f'mandelbrot_{width}x{height}.png'
print(f'Writing image to {output_file}')
plt.imsave(output_file, result)
Try running this script under rx:
$ rx python mandelbrot.py
Writing image to mandelbrot_400x400.png
Changed:
mandelbrot_400x400.png
rx does several things here: it syncs your local project state to the remote machine, runs your script (python mandelbrot.py), and finally syncs the output file (mandelbrot_400x400.png) back to your local machine. If you take a look at your local directory, you can see the output file:
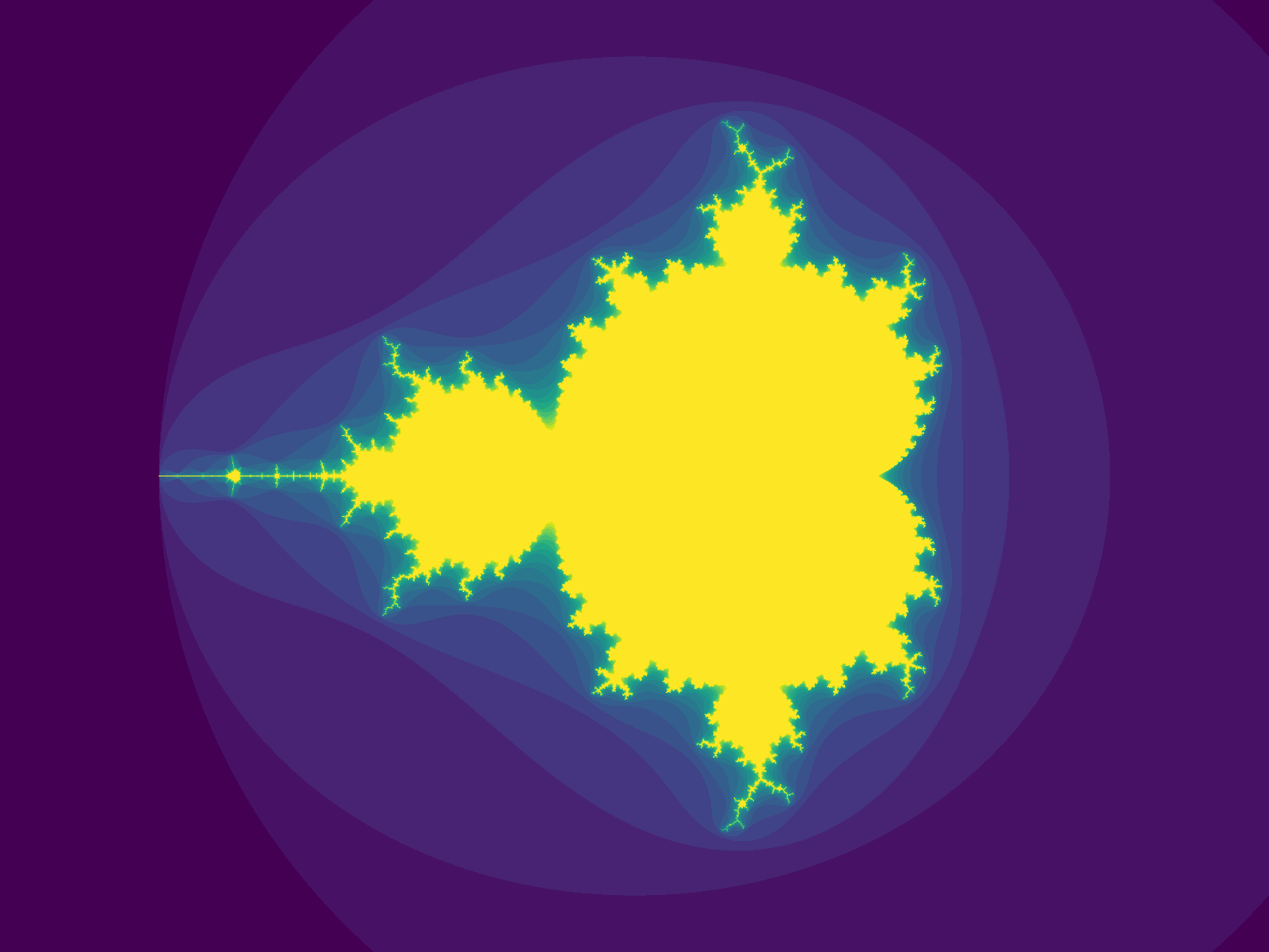
However, usually running code isn't one-and-done: you want to want to have a tight write/run loop. rx gets out of your way for this: e.g., try changing the height and width parameters locally:
height = 200
width = 400
Now rerun:
rx python mandelbrot.py
Changed:
mandelbrot_400x200.png
The remote machine picks up on your changes immediately and the new output is synced back to your local machine. This allows you to quickly iterate locally while still using powerful and/or annoying libraries for execution.
To learn more about rx check out the documentation (and subscribe to this blog!).
Subscribe to my newsletter
Read articles from k directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
