Integrating LlamaIndex and Qdrant Similarity Search for Patient Record Retrieval
 Akriti Upadhyay
Akriti Upadhyay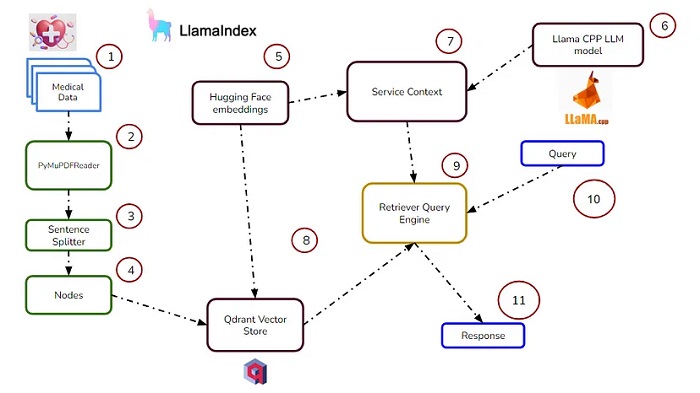
Introduction
The medical field is currently experiencing a remarkable surge in data, a result of the progress in medical technologies, digital health records (EHR), and wearable health devices. The ability to effectively manage and analyze this intricate and varied data is vital for providing customized healthcare, advancing medical research, and enhancing patient health outcomes. Vector databases, which are specifically tailored for the efficient handling and storage of multi-dimensional data, are gaining recognition as an effective tool for a range of healthcare uses.
For example, currently, past patient record data is rarely leveraged by medical professionals in real-time, even though they are a treasure trove of information and can assist in diagnosis. What if we could build systems where doctors, nurses and caregivers could quickly access past patient records using just natural language inputs? What if historical test results could help generate recommendations for new treatment options?
This is the potential of AI in healthcare. From personalized diagnostics to targeted therapies, healthcare is on the cusp of becoming a whole lot smarter. In this article, I will demonstrate the capabilities and potential applications of vector databases in the healthcare sector.
Why Vector Search and LLMs?
Vector Search enables rapid exploration of large datasets by transforming data into vectors within a high-dimensional space, where similar items are clustered closely. This approach facilitates efficient retrieval of relevant information, even from vast datasets. LLMs, on the other hand, are AI models trained on diverse internet texts, capable of comprehending and generating human-like text based on inputs.
When combined, Vector Search and LLMs streamline the storage and search process for patient records. Each record undergoes embedding and converts it into a vector representing its semantic meaning, which is then stored in a database. During retrieval, a doctor inputs a search query, also converted into a vector, and the Vector Search scans the database to locate records closest to the query vector, which enables semantic search based on meaning rather than exact keywords.
Subsequently, retrieved records are processed through an LLM, which generates a human-readable summary highlighting the most relevant information for the doctor. This integration empowers doctors to efficiently access and interpret patient records, which facilitates better-informed decisions and personalized care. Ultimately, this approach enhances patient outcomes by enabling healthcare professionals to provide tailored recommendations based on comprehensive data analysis.
Let’s see how this is going to work with the help of the Retrieval Augmented Generation (RAG) technique incorporated with LlamaIndex and Qdrant Vector DB.
RAG Architecture
Retrieval Augmented Generation (RAG) enhances the effectiveness of large language model applications by incorporating custom data. By retrieving relevant data or documents related to a query or task, RAG provides context for LLMs, which improves their accuracy and relevance.
The challenges addressed by RAG include LLMs' limited knowledge, which is beyond their training data, and the necessity for AI applications to leverage custom data for specific responses. RAG tackles these issues by integrating external data into the LLM's prompt, which allows it to generate more relevant and accurate responses without the need for extensive retraining or fine-tuning.
RAG benefits by reducing inaccuracies or hallucinations in LLM outputs by delivering domain-specific and relevant answers, and by offering an efficient and cost-effective solution for customizing LLMs with external data.
Incorporating RAG with LlamaIndex
LlamaIndex is a fantastic tool in the domain of Large Language Model Orchestration and Deployment and particularly focuses on Data Storage and Management. Its standout features include Data Agents, which execute actions based on natural language inputs instead of generating responses, and it can deliver structured results by leveraging LLMs.
Moreover, LlamaIndex offers composability by allowing the composition of indexes from other indexes. It also has seamless integration with existing technological platforms like LangChain, Flask, and Docker, and customization options such as seeding tree construction with custom summary prompts.
Qdrant DB: A High-Performance Vector Similarity Search Technology
Qdrant acts both as a vector database and similarity search engine and has a cloud-hosted platform that helps find the nearest high-dimensional vectors efficiently. It harnesses embeddings or neural network encoders to help developers build comprehensive applications that involve tasks like matching, searching, recommending, and beyond. It also utilizes a unique custom adaptation of the HNSW algorithm for Approximate Nearest Neighbor Search. It allows additional payload associated with vectors and enables filtering results based on payload values.
Qdrant supports a wide array of data types and query conditions for vector payloads by encompassing string matching, numerical ranges, geo-locations, and more. It is built to be cloud-native and horizontally scalable. Qdrant maximizes resource utilization with dynamic query planning and payload data indexing which is implemented entirely in Rust language.
The HNSW Algorithm
There are many algorithms for approximate nearest neighbor search, such as locality-sensitive hashing and product quantization, which have demonstrated superior performance when handling high-dimensional datasets.
However, these algorithms, often referred to as proximity graph-ANN algorithms, suffer from significant performance degradation while dealing with low-dimensional or clustered data.
In response to this challenge, the HNSW algorithm has been developed as a fully graph-based incremental approximate nearest neighbor solution.
The HNSW algorithm builds upon the hierarchical graph structure of the NSW algorithm. While the NSW algorithm struggles with high-dimensional data, its hierarchical counterpart excels in this domain by offering optimal performance. The core concept of the HNSW algorithm involves organizing links based on their length scales across multiple layers. This results in an incremental multi-layer structure that comprises hierarchical sets of proximity graphs, each representing nested subsets of the stored elements within the NSW. The layer in which an element resides is chosen randomly, which follows an exponentially decaying probability distribution.
Building Medical Search System with LlamaIndex
To get started with utilizing RAG for building a medical search system, let’s create a synthetic dataset first.
Generating Synthetic Patient Data
As this is going to be synthetic data, let’s install a dependency named ‘Faker’.
!pip install -q faker |
Create a CSV synthetic dataset.
import random |
After the synthetic dataset is created, it will print that it’s created, and you can see it in your directory.
Synthetic medical history dataset created and saved to 'medical_history_dataset.csv' |
Let’s see what our data looks like!
import pandas as pd |
The data looks fine; let’s convert it into a PDF format. While we could use the CSV format as is, PDF is the format in which many documents are stored in legacy systems – so using PDF as a base is a good way to build it for real-life scenarios.
We will load the PDF data using LlamaIndex SimpleDirectoryReader.
To convert the CSV dataset into a PDF document, install the following dependency.
!pip install -q reportlab |
from reportlab.lib.pagesizes import letter |
Make a directory and move the output pdf document into the directory.
import os |
Now that the dataset is ready, let’s move to building an RAG using this dataset. Install the important dependencies.
!pip install -q llama-index transformers |
Load the data into SimpleDirectoryReader.
from llama_index import SimpleDirectoryReader |
Using the Sentence splitter, split the documents into small chunks. Maintain the relationship between the Source document index, so that it helps in injecting document metadata.
from llama_index.node_parser.text import SentenceSplitter |
Now, manually construct nodes from text chunks.
from llama_index.schema import TextNode |
Now generate embeddings for each node using the Hugging Face embeddings model.
from llama_index.embeddings import HuggingFaceEmbedding |
Now, it’s time to build a model using Llama CPP. Here, we’ll use the GGUF Llama 2 13B model. Using Llama CPP, we’ll download the model with the help of the model URL.
from llama_index.llms import LlamaCPP |
Let’s define Service context. It consists of the LLM model as well as the embedding model.
from llama_index import ServiceContext |
Now, create a vector store collection using Qdrant DB, and create a storage context for this vector store.
import qdrant_client |
Now, create an index of the vector store where the service context, storage context, and the documents are stored.
index = VectorStoreIndex.from_documents( |
Add the created node into the vector store.
vector_store.add(nodes) |
To build a retrieval pipeline, generate a query embedding using a query string.
query_str = "Can you tell me about the key concepts for safety finetuning" |
Then, construct a Vector Store query and query the vector database.
from llama_index.vector_stores import VectorStoreQuery |
You’ll get the following results:
Patient_ID: 58b70a59-eb30-4caa-b4b5-7871321515dd, Name: Kimberly Brown, Age: |
Now, parse the results into a set of nodes.
from llama_index.schema import NodeWithScore |
Then, put them into a retriever.
from llama_index import QueryBundle |
Create a Retriever Query Engine, and plug the above into it to synthesize the response.
from llama_index.query_engine import RetrieverQueryEngine |
Now, it’s time to query the Retriever Query Engine and see the response.
query_str = "Write prescription for Diabetes" |
It’ll take a significant amount of time; be patient, and you will get the response:
Metformin, Dosage: 3 pills, Duration: 12 days |
Let’s see the source node of this response.
print(response.source_nodes[0].get_content()) |
Following is the Source Node:
Patient_ID: ea9121cf-22d3-4053-9597-32816a087d6b, Name: Tracy Mendez, Age: |
Note: The medical dataset used here is synthetic and fake. It has no relation to real medicine dosage or duration.
Conclusion
Leveraging the RAG architecture with tools like LlamaIndex, Llama CPP, and Qdrant Vector Store has been a fascinating journey. Through the utilization of Qdrant's sophisticated HNSW algorithm, searching through patients' medical histories and records has become effortless and rapid. This integration highlights the potential of innovative technologies to enhance healthcare processes, which can ultimately lead to improved patient care and outcomes.
This article was originally posted here: https://medium.com/@akriti.upadhyay/integrating-llamaindex-and-qdrant-similarity-search-for-patient-record-retrieval-7090e77b971e
Thanks for reading!
Subscribe to my newsletter
Read articles from Akriti Upadhyay directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
