Text Extraction for Information Retrieval using LLMsherpa, Neo4j, and LangChain
 Asmaa Hadir
Asmaa Hadir
The Challenge: LLMs and Large Texts
Imagine being tasked with piecing together a puzzle, but you're only allowed to see a few pieces at a time without ever seeing the whole picture. That's the dilemma facing today's most advanced computer programs in understanding human language, known as natural language processing (NLP). These programs, called large language models (LLMs), are at the forefront of analyzing complex documents like PDFs, yet they stumble when confronted with lengthy texts. The challenge? These intelligent systems are designed to digest only snippets of text at once, akin to trying to grasp a novel's narrative by reading a few sentences at a time. The result is a fragmented understanding of the content, leaving the big picture - and often crucial details - obscured.
This limitation not only complicates the task of analyzing large documents but also restricts the potential applications of NLP in fields where comprehensive document analysis is essential. From creating knowledge graphs that enhance question-answering systems to developing vector stores for text chunk embeddings, and automating the summarization of extensive materials, the ability to fully understand and process large documents could dramatically improve efficiency and insight.
LLMSherpa
Enter LLMsherpa, an open source tool designed by Nlmatics to navigate this very challenge and assist developers in building LLM projects. Far from a mere text slicer, LLMsherpa acts as an astute guide for LLMs, enabling them to traverse and comprehend the entirety of large documents without losing their way. By intelligently segmenting texts in a manner that maintains narrative coherence and thematic integrity, it ensures that every piece of the puzzle is placed correctly, allowing for a complete and nuanced understanding. With LLMsherpa, we're not just reading more text; we're unlocking the true potential of language models to grasp the full story, revolutionizing how we interact with the vast seas of digital information. You could read more about LLMsherpa's approach and find its documentation on its Github repository
In this article, we'll dive directly into a hands-on demonstration, using LLMsherpa to perform smart chunking on the Retrieval Augmented Generation (RAG) paper by Lewis et al. and use the extracted data to build a knowledge graph database on Neo4j for intuitive retrieval. If you are not familiar with graph databases, I would highly recommend referring to Victoria Lo 's brilliant intro to Neo4j and graph databases. Briefly put: a graph database is a database which represents its data as (you guessed it) a graph! this provides the user with a more visualized and intuitive tool for storing and retrieving information while maintaining the database's entities' relationships and properties.
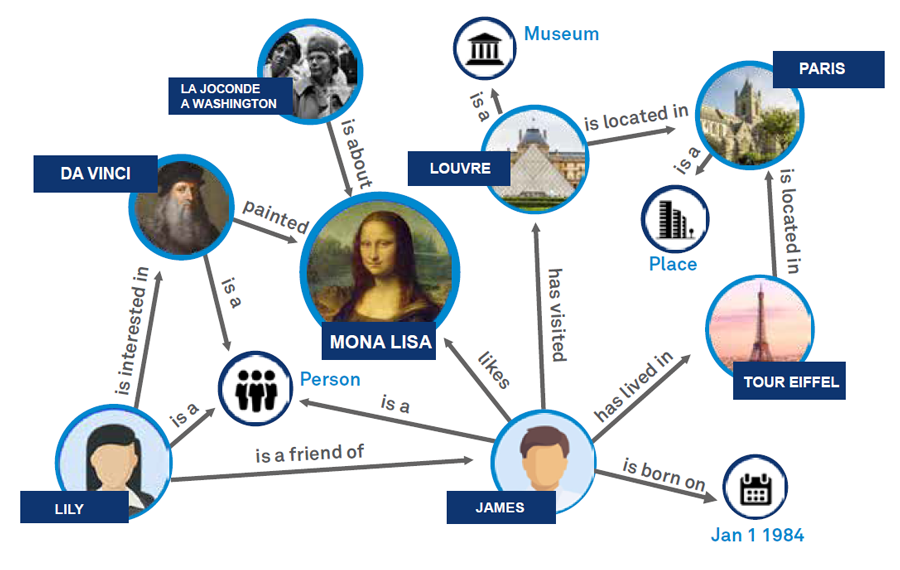
The document we have chosen is an ideal choice because it includes a mix of elements common in real-world PDF document mining projects: images, tables, and a hierarchical structure of sections and subsections. This variety presents just the right level of complexity for our demonstration.


Tutorial
Installation
To install LLMSherpa, you can run the following command:
pip install llmsherpa
Reading the Document
The core of LLMsherpa's smart chunking is how it represents the input PDF documents. These documents are read as a layout tree which fully preserves the natural section-subsection hierarchy of the document. To read the PDF at hand we use the layoutPDFReader submodule inside the file_reader module.
from llmsherpa.readers import LayoutPDFReader
llmsherpa_api_url = "https://readers.llmsherpa.com/api/document/developer/parseDocument?renderFormat=all"
pdf_url = "/assets/RAG_paper.pdf"
pdf_reader = LayoutPDFReader(llmsherpa_api_url)
doc = pdf_reader.read_pdf(pdf_url)
The LayoutPDFReader returned is an object that reads and understands the content and hierarchical layout of the document's sections and their different components. The LayoutPDFReader must take LLMsherpa API url as an argument. We can then use this reader object to read our document and in return we get a llmsherpa.readers.layout_reader.Document object. This returned Document object is the root node of the layout tree and each Document object is a tree of blocks. What is a block you ask? a block can be a paragraph, a list item, a table, a section header or any other element inside our document. A block is basically a node in the layout tree, and we can view all of the document's blocks by using the Document.json property:
blocks = doc.json
print(blocks)
# Output
[{'block_class': 'cls_0',
'block_idx': 0,
'level': 0,
'page_idx': 0,
'sentences': ['Retrieval-Augmented Generation for Knowledge-Intensive NLP Tasks Patrick Lewis†‡, Ethan Perez?, Aleksandra Piktus†, Fabio Petroni†, Vladimir Karpukhin†, Naman Goyal†, Heinrich Küttler†, Mike Lewis†, Wen-tau Yih†, Tim Rocktäschel†‡, Sebastian Riedel†‡, Douwe Kiela†'],
'tag': 'para'},
{'block_class': 'cls_3',
'block_idx': 1,
'level': 0,
'page_idx': 0,
'sentences': ['†Facebook AI Research; ‡University College London; ?New York University; plewis@fb.com'],
'tag': 'para'},
{'block_class': 'cls_6',
'block_idx': 2,
'level': 0,
'page_idx': 0,
'sentences': ['Abstract Large pre-trained language models have been shown to store factual knowledge in their parameters, and achieve state-of-the-art results when fine-tuned on downstream NLP tasks.',
'However, their ability to access and precisely manipulate knowledge is still limited, and hence on knowledge-intensive tasks, their performance lags behind task-specific architectures.',
'Additionally, providing provenance for their decisions and updating their world knowledge remain open research problems.',
'Pretrained models with a differentiable access mechanism to explicit non-parametric memory have so far been only investigated for extractive downstream tasks.',
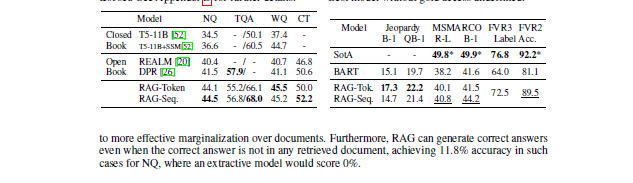
'We explore a general-purpose fine-tuning recipe for retrieval-augmented generation (RAG) — models which combine pre-trained parametric and non-parametric memory for language generation.',...]}]
Each block in the blocks list has multiple properties including sentences which contains the text content of that block and tag which indicates the type of the block: para for paragraph, header for a header title, table for a table etc.
As you can see we have access to a plethora of rich information about the content of our text through these blocks. However, they are too granular for our scope of text extraction. We do not want to grab the content of our document paragraph by paragraph or header by header, we rather want to have access to whole sections or chunks of the document. Luckily we have access to both:
chunks(): this method retrieves all of the chunks in the document's block.sections(): this method on the other hand retrieves all of the sections present in the document with other info such as a section's title, page number tec.
From my experience, the chunks() is most useful if you are looking to efficiently split text for embeddings generation to be stored in a vector store. On the other hand, if you are intending to store your document in a knowledge graph database, then sections() method may be the ideal choice to extract sections and their subsections, exactly what we will do right now. But first, let us briefly check the results of the chunks method.
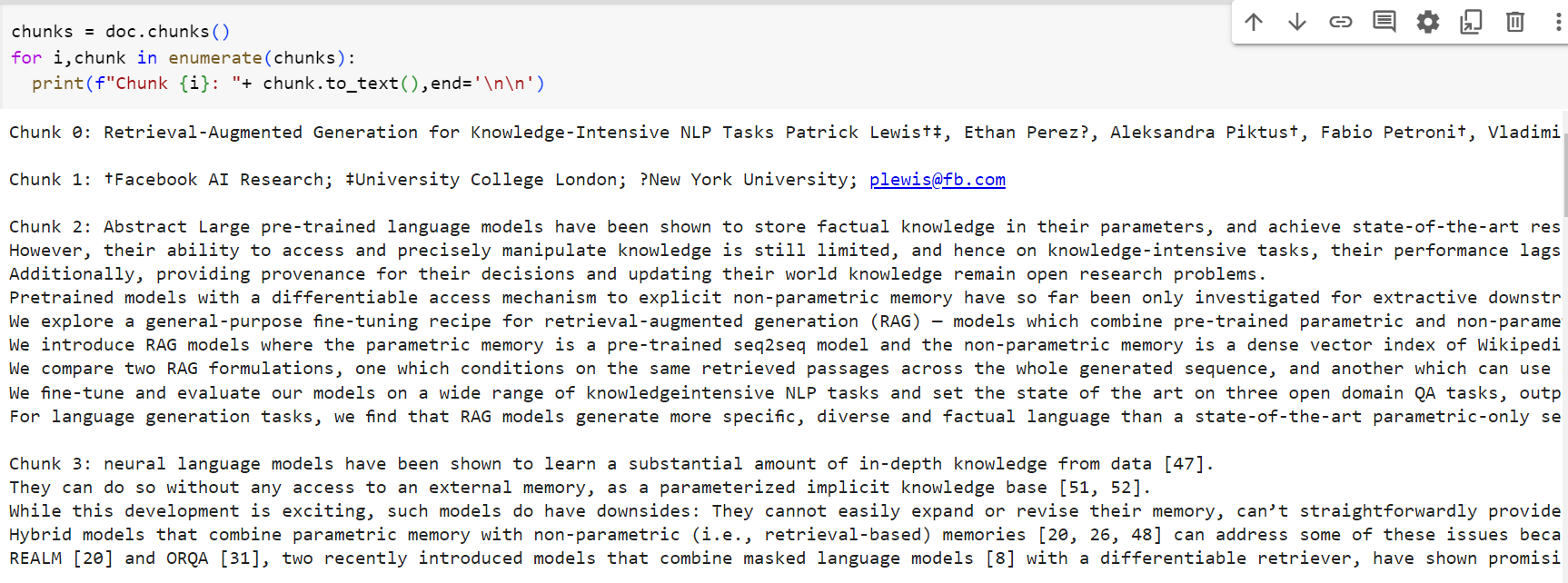
By running the above code, we get the chunks into which LLMsherpa divides the document. These chunks are Paragraph objects. If you run the code over your document you will notice that this chunking achieves higher quality than many other chunking tools such that each chunk doesn't abruptly end at a random position in the paragraph which will ultimately help you achieve higher accuracy in information retrieval later on.
Extracting The Text Sections
Let us create a script which will allow us to build a dictionary with our document's sections and their info:
import uuid
def get_file_sections(doc):
sections = []
sections_lst = doc.sections() # access the document's sections
for section in sections_lst:
section_dict = {}
section_txt = section.to_text(include_children=True) # get the text content of the section, include_children flag tells the api that we want to include the children sections as well
section_title = section.title
section_dict = {"title":section_title,"text":section_txt,"id":uuid.uuid4().int}
sections.append(section_dict)
return sections
sections = get_file_sections(doc)
The above code uses the sections properties and methods to grab each section's title and text content. The returned value is a list of dictionaries representing all of the document's sections. The results look like this:

Now that we grabbed these sections we need to slightly modify our dictionary by adding a subsection field to each parent section. This field will contain a list of child sections. To do this, we notice that subsections' titles are of the format "x.ytitle" where X corresponds to the major section's number, so we can grab each section with a title of that format and append it to the last major section's subsection field until we encounter a new major section and so on. Note that since our sections is a list then it preserves the order of sections extracted from the documents, so we do not have to worry about randomly and mistakenly assigning a subsection to a section which is not its parent section in the actual doc.
def add_hierarchy(sections):
# Prepare a mapping from titles to section dictionaries
title_to_section = {section['title']: section for section in sections}
# Initialize an empty list for subsections in each dictionary
for section in sections:
section['subsections'] = []
# Populate subsections
for section in sections:
title_parts = section['title'].split(' ')
if '.' in title_parts[0]: # It's a subsection
major_section_number = title_parts[0].split('.')[0]
# Find the corresponding major section and add this subsection
for major_section_title, major_section in title_to_section.items():
if major_section_title.startswith(major_section_number + ' '): # Matching major section
major_section['subsections'].append(section)
break # Found the matching major section, no need to continue
# Apply the function
add_hierarchy(sections)
The output looks like the following:


This looks decent enough! now we can proceed with inserting this data to our Neo4j graph. First we define the cypher queries which we will run to create the needed nodes and relationships:
create_section_node = """
MERGE (n:section {title: $section_title, text: $section_text, id: $section_id})
"""
create_relationship = """
MATCH (s1:section {id: $id1}), (s2:section {id: $id2})
MERGE (s1)-[r:CONTAINS]->(s2)
"""
creat_section_node and create_relationship both contain Cypher queries. Cypher is the language that we use to run queries over graph databases like Neo4j.
First, we install the neo4j library:
pip install neo4j
Next, we will use Neo4j`s Python driver to connect to our database and define a function to run our queries.
from neo4j import GraphDatabase
import os
from dotenv import load_dotenv
load_dotenv()
url = os.getnev['NEO4J_URI']
username = os.getnev['NEO4J_USERNAME']
password = os.getnev['NEO4J_PASSWORD']
driver = GraphDatabase.driver(url, auth=(username, password))
def execute_query(query, parameters=None):
with driver.session() as session:
session.run(query, parameters)
url, username , and password correspond to the credentials you receive after creating a new Neo4j Auradb instance. Follow this guide to sign up and create your first Neo4j Auradb instance. You can also create your Neo4j DBSM locally on Neo4j Desktop.
Finally, we define a function to iterate over the sections and create their nodes, their subsections' nodes and link them with a relationship of type Contains such that a section contains its subsection(s):
def insert_to_graph(sections):
for i,section in enumerate(sections):
# first, insert high level section
execute_query(create_section_node,{'section_title': section['title'],
'section_text':section['text'],
'section_id':str(section['id'])})
# then if high level section has subsections, create their nodes and their relationship to their parent section node
if section['subsections']:
for subsection in section['subsections']:
execute_query(create_section_node,{
'section_title': subsection['title'],
'section_text':subsection['text'],
'section_id':str(subsection['id'])
})
execute_query(create_relationship,{
'id1':str(section['id']),
'id2':str(subsection['id'])
})
print(f"inserted section number {i}")
And simply run it to insert the nodes and relationships:
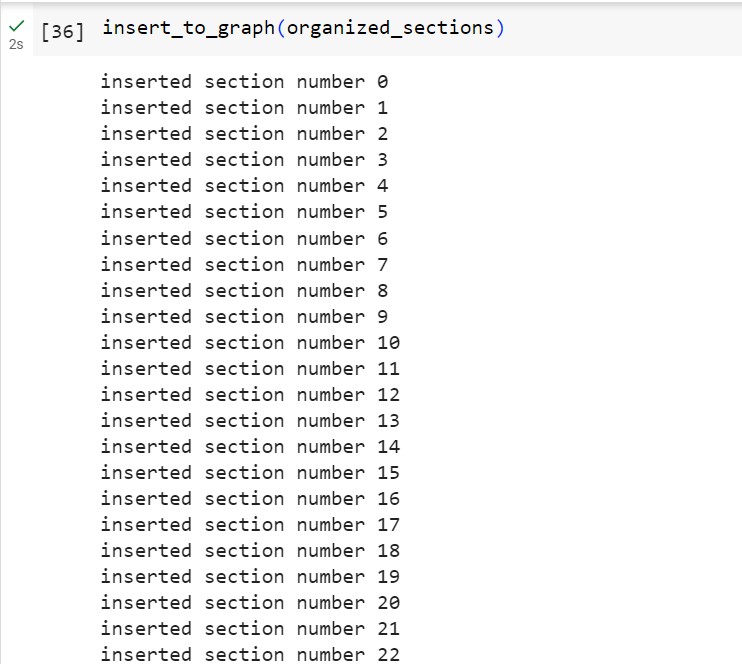
And voila! our graph database contains our document's sections and subsections:
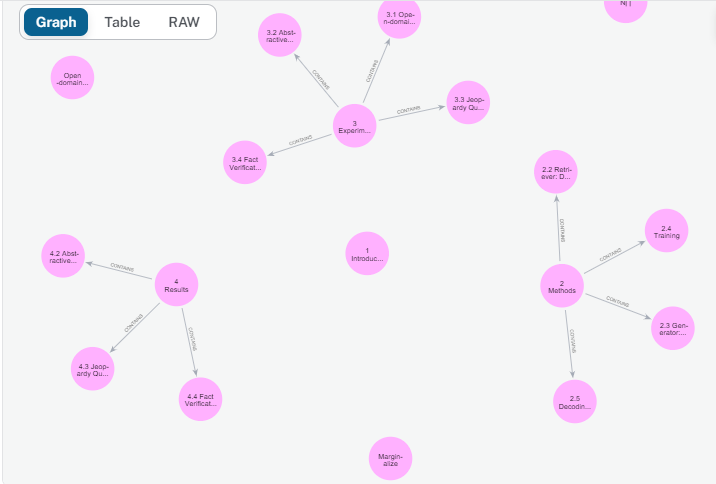
Each pink circle is a node in our graph connected to its subsections by an outgoing arrow indicating a section-Contains-section relationship. We also notice that there are some missing sections such as the 2.1 and 4.1 sections, and if we inspect the document's blocks Json object we will notice that they are not detected at a Header element, which brings us to one of our last points that text extraction APIs may not be 100% accurate but they definitely help us get the majority of the work done. It is very important to closely inspect our data, track the unprocessed data elements, and identify what made me go under the radar then write custom text extraction script accordingly.
Just like that we went from a PDF document to building a rich graph knowledge base that we can use for many powerful applications including Q&A. Luckily, LangChain provides an intuitive API to perform Q&A over Neo4j databases.
Since we will be using LangChain's ChatOpenAI() as our chat model, make sure that you grab your OpenAI API key and store it as an environment variable as OPENAI_API_KEY (you can also pass it directly to the chat model using the openai_api_key keyword argument).
from langchain.prompts.prompt import PromptTemplate
from langchain_community.graphs import Neo4jGraph
from langchain.chains import GraphCypherQAChain
from langchain_openai.chat_models import ChatOpenAI
# connect to db using Langchain's Neo4jGraph integration
graph = Neo4jGraph(
url=url,
username=username,
password=password
)
# if schema changes refresh it
graph.refresh_schema()
chain = GraphCypherQAChain.from_llm(
ChatOpenAI(temperature=0,model="gpt-3.5-turbo",openai_api_key=OPENAI_API_KEY),
graph=graph, verbose=True,
validate_cypher=True
)
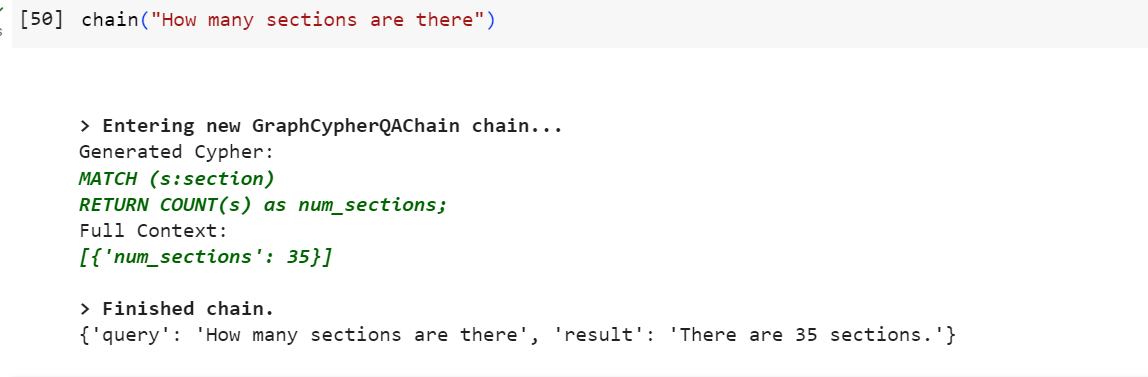
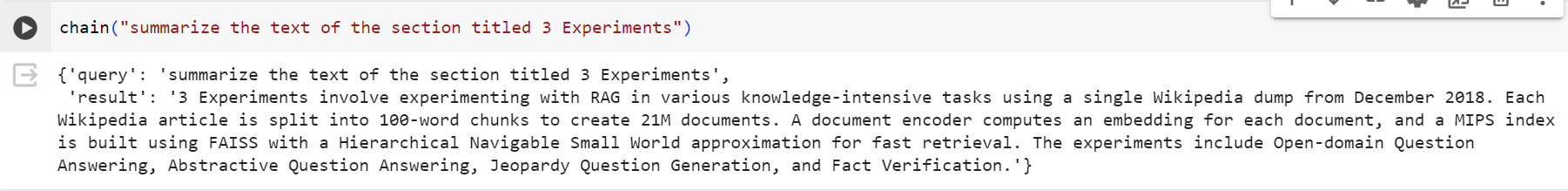
We are ready to ask some questions!
Tables Extraction
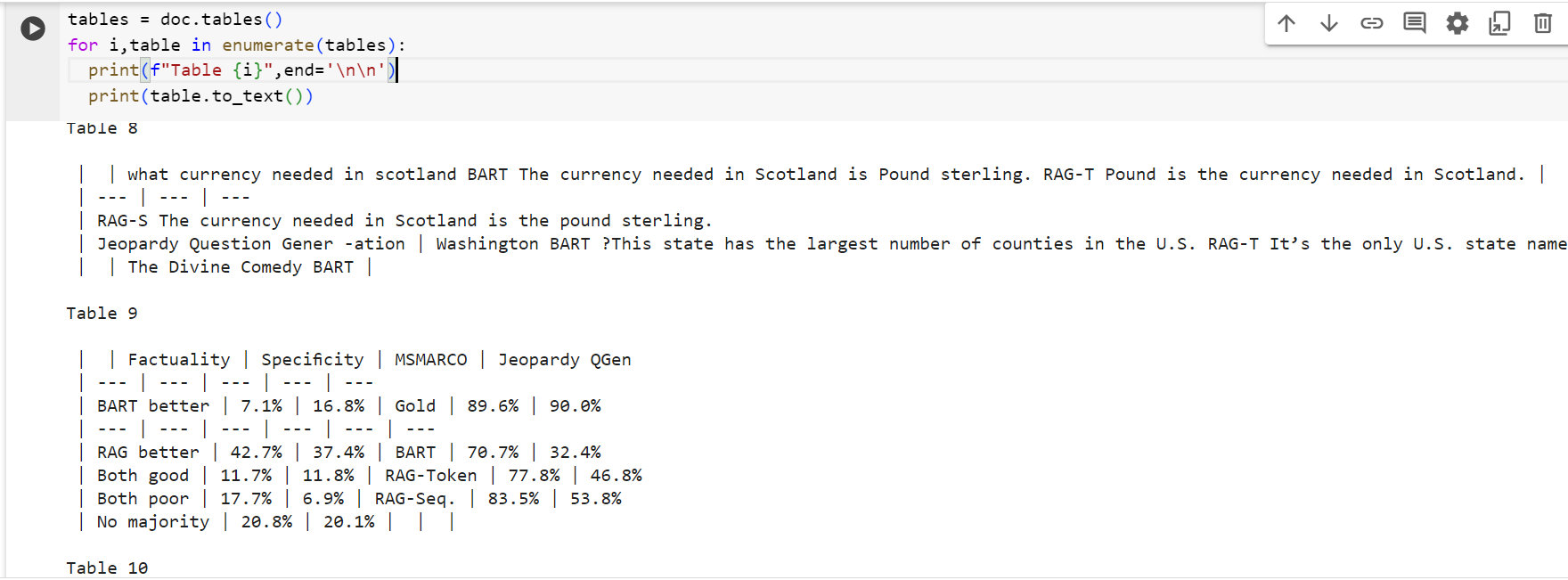
You can also extract tables from any pdf document using the tables() and then by applying the to_text() method, you can obtain a simple textual representation of each of these tables. This means that you can expand your Large Language Model's ability to answer questions about tables' data etc.
tables = doc.tables()
for i,table in enumerate(tables):
print(f"Table {i}",end='\n\n')
print(table.to_text())
The above code displays each table from the tables list.
Real-World Challenges in Text Extraction for NLP Engineers
In the real world, NLP and machine learning engineers often encounter documents that defy simple analysis: legal contracts with dense and arcane language, scientific papers rich in specialized terminology and complex diagrams, and historical texts with outdated language or poor digitization quality. For instance, extracting usable data from a 19th-century legal document involves not just understanding the archaic language but also dealing with scanned images where text may be faded or obscured. Similarly, analyzing modern scientific literature requires not only parsing the text but also understanding and extracting information from charts, graphs, and tables that are critical to the document's meaning. These examples highlight the multifaceted challenges in text extraction, from dealing with non-standard language and document damage, to interpreting non-textual information—a testament to the sophistication required in NLP tools and the ingenuity demanded of their developers.
Conclusion
In summary, this article has unveiled how large language models (LLMs), specifically through the lens of LLMSherpa, are revolutionizing our ability to handle and analyze large documents in natural language processing (NLP). By breaking down the barrier of limited text input size, LLMSherpa offers a smarter approach to digesting extensive texts, ensuring that the essence and structure of documents are preserved and understood in their entirety. Through a hands-on demonstration with the Retrieval Augmented Generation (RAG) paper and the use of Neo4j for knowledge graph construction, we've seen how these advanced tools can be applied to real-world data for more intuitive information retrieval and analysis.
As we close, consider the vast potential of combining LLMs with innovative solutions like LLMSherpa: How might these advancements further transform our interaction with the ever-growing expanse of digital information? The future of NLP and information processing is bright, with endless possibilities for exploration and innovation.
Subscribe to my newsletter
Read articles from Asmaa Hadir directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by

Asmaa Hadir
Asmaa Hadir
I am a Natural Language Processing Engineer with experience and expertise in building LLM-backed applications and chatbots. Holder of a Bachelor's degree in Computer Science from the American University of Beirut.