Adding Documentation support for MedImages.jl and MedEye3D.jl packages
 Divyansh Goyal
Divyansh Goyal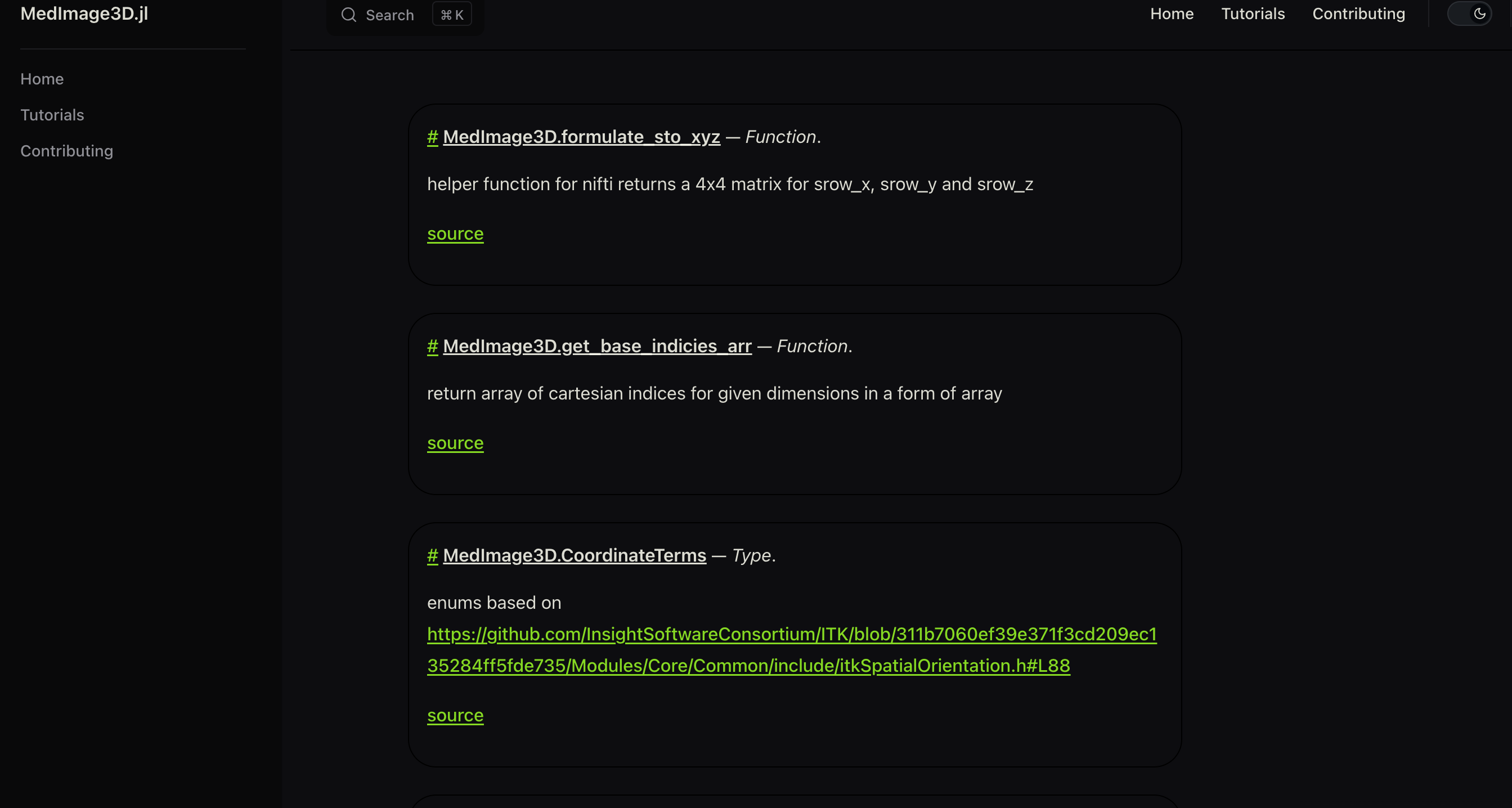
In order to expand the medical imaging scope within the julia language ecosystem, MedImage3D.jl package was introduced to handle BIDS format medical imaging data, based on the ideas from SimpleITK / ITK -InsightToolkit.
MedImage3D.jl package introduces a data structure that handles data from "nifti", "dicom" and "HDF5" file format sources, and produces an array of objects (based on the file instances given) with fields for holding spatial metadata such orientation , origin , spacing and direction, crucial for medical imaging pipelines to ensure correct image registrations within visualization utilities.
Navigating through the blunders to get to a working solution
Recently the entire JuliaHealth community have been migrating from the legacy Documenter.jl code for maintaining documentation to a more vibrant and modern documentation curator DocumenterVitepress.jl. Both Documenter and DocumenterVitepress are put in place to ensure the deployment and management of package docstrings.
Workflow for establishing a successful DocumenterVitepress documentation deployment
In order to create documentation for a package, we first need to ensure that the pakage is present in the official Julia Registry, in order to provide a callable interface from the Julia REPL. If the package is currently under development and not yet been released, there is a workaround which is explained in the next section.
Let's start by understanding the directory structure setup for documentervitepress,
Getting Started with DocumenterVitepress
The documentation of documentervitepress mentions the directory structure of the project being,
docs
|__Project.toml
|__make.jl
|__package.json
|__src
|__assets
|__image_one.png
|__index.md
|__otherfiles.md
the above directory structure needs to be put in place in order to deploy a documentation for a package. Within the root of a package a "docs" folder is created with a Project.toml. This Project.toml file is independent from the package's Project.toml files present inside of the root of the directory.
Essentially, we are trying to setup an independent project environment inside of this docs directory in order to produce documentation. But the dependencies inside of the Project.toml file should be a part of the package's project.toml file with correction compatibility version strings.
Project.toml file for docs
[deps]
Documenter = "e30172f5-a6a5-5a46-863b-614d45cd2de4"
DocumenterVitepress = "4710194d-e776-4893-9690-8d956a29c365"
The above toml file declares the environment's dependencies being Documenter.jl and DocumenterVitepress.jl followed by their Universally Unique iD strings or UUIDs (just navigate to each of these packages github repositories and find the UUID in the top of their own Project.toml files).
Project.toml present in the root of the package
name = "MedImage3D"
uuid = "fbff095b-7891-4fc4-b5e5-3cd99b9dd225"
authors = ["Jakub Mitura <jakub.mitura@gmail.com>"]
version = "0.1.0"
[deps]
Accessors = "7d9f7c33-5ae7-4f3b-8dc6-eff91059b697"
BinaryBuilder = "12aac903-9f7c-5d81-afc2-d9565ea332ae"
ColorTypes = "3da002f7-5984-5a60-b8a6-cbb66c0b333f"
Combinatorics = "861a8166-3701-5b0c-9a16-15d98fcdc6aa"
Conda = "8f4d0f93-b110-5947-807f-2305c1781a2d"
CoordinateTransformations = "150eb455-5306-5404-9cee-2592286d6298"
DICOM = "a26e6606-dd52-5f6a-a97f-4f611373d757"
Dates = "ade2ca70-3891-5945-98fb-dc099432e06a"
Documenter = "e30172f5-a6a5-5a46-863b-614d45cd2de4"
DocumenterVitepress = "4710194d-e776-4893-9690-8d956a29c365"
Dictionaries = "85a47980-9c8c-11e8-2b9f-f7ca1fa99fb4"
HDF5 = "f67ccb44-e63f-5c2f-98bd-6dc0ccc4ba2f"
ImageTransformations = "02fcd773-0e25-5acc-982a-7f6622650795"
Interpolations = "a98d9a8b-a2ab-59e6-89dd-64a1c18fca59"
JLD = "4138dd39-2aa7-5051-a626-17a0bb65d9c8"
JSON = "682c06a0-de6a-54ab-a142-c8b1cf79cde6"
LinearAlgebra = "37e2e46d-f89d-539d-b4ee-838fcccc9c8e"
NIfTI = "a3a9e032-41b5-5fc4-967a-a6b7a19844d3"
Parameters = "d96e819e-fc66-5662-9728-84c9c7592b0a"
Pkg="44cfe95a-1eb2-52ea-b672-e2afdf69b78f"
PyCall = "438e738f-606a-5dbb-bf0a-cddfbfd45ab0"
PythonCall = "6099a3de-0909-46bc-b1f4-468b9a2dfc0d"
Rotations = "6038ab10-8711-5258-84ad-4b1120ba62dc"
Setfield = "efcf1570-3423-57d1-acb7-fd33fddbac46"
StaticArrays = "90137ffa-7385-5640-81b9-e52037218182"
Statistics = "10745b16-79ce-11e8-11f9-7d13ad32a3b2"
[compat]
Accessors = "0.1.36"
Documenter = "1"
DocumenterVitepress = "0.0.19, 0.0.20"
Above is the Project.toml for MedImage3D.jl pkg declaring a bunch of package dependencies along with the presence of Documenter and DocumenterVitepress under [deps]. Further in order to make sure that there are no version compatibility issues with DocumenterVitepress, we are declaring the version compatibility strings for Documenter with DocumenterVitepress (DocumenterVitepress requires Documenter version 1.0.0 and above).
Now, the most crucial part of the process is the creation of a make.jl file inside of docs directory, which is going to manage the package's documentation, urls, deployment and content from the src.
Here Assuming that the MedImage3D pkg is in the registry (currently's it is not).
using MedImage3D
using Documenter
using DocumenterVitepress
DocMeta.setdocmeta!(MedImage3D, :DocTestSetup, :(using MedImage3D); recursive=true)
makedocs(;
modules = [MedImage3D],
repo = Remotes.GitHub("divital-coder", "MedImage.jl"),
authors = "Jakub-Mitura <jakubmitura14@gmail.com>, Divyansh-Goyal <divital2004@gmail.com> and contributors",
sitename = "MedImage3D.jl",
format = DocumenterVitepress.MarkdownVitepress(
repo="https://github.com/JuliaHealth/MedImage.jl",
),
pages = [
"Home" => "index.md",
"Tutorials" => "tutorials.md",
"Contributing" => "contributing.md"
],
)
deploydocs(;
repo = "github.com/divital-coder/MedImage.jl",
target = "build", # this is where Vitepress stores its output
devbranch = "main",
branch = "gh-pages",
push_preview = true
)
The above make.jl file uses functions from the package, documenter and documentervitepress to produce the documentation pages and deployment with preferred formatting and endpoints.
The setdocmeta sets metadata for documentation, and other functions are necessary for documentation deployment and page content. For more information take a look at the following :
Migration from Documenter to DocumenterVitepress
A few things to note here are, since the branch for deployment is set to "gh-pages" the repository must have a branch named "gh-pages", if there is not create one.
Further make sure to correctly identify between your forked and original upstream repository url. Here since my Fork of MedImage had changes with a new branch and gh-pages, i had to edit my github username in the url so to make sure the local deployment takes place on my fork of the repository. This is only the case, when the upstream doesn't have a branch named "gh-pages" yet.
Another thing to note here is about the mention of the dev-branch, make sure that the name of your dev-branch matches the name on the makefile.jl.
Creating documentation content under the src directory within "docs".
The documentation content needs to be laid out in makdown format with a required index.md file with the function docstrings of the package, and additional markdown files as per the need of the documentation for the package. This should be in sync with your revisions within the make.jl file "pages" variable.
The index.md should contain all the function names your package, (doc strings) as per the following example format :
```@docs
MedImage3D.formulate_sto_xyz
MedImage3D.get_base_indicies_arr
MedImage3D.CoordinateTerms
MedImage3D.interpolate_my
MedImage3D.unique_series_id_within_dicom_files
MedImage3D.formulate_xform_string
MedImage3D.resample_to_image
MedImage3D.calculate_inverse_44_matrix
MedImage3D.formulate_string
MedImage3D.interpolate_point
MedImage3D.Interpolator_enum
MedImage3D.scale_mi
MedImage3D.Mode_mi
MedImage3D.scale
MedImage3D.Image_type
MedImage3D.cast_to_array_b_type
MedImage3D.Image_subtype
MedImage3D.string_intent
MedImage3D.formulate_timing_scale_for_xyzt_time
MedImage3D.set_direction_for_nifti_file
MedImage3D.get_pixel_data
MedImage3D.set_spacing_for_nifti_files
MedImage3D.set_origin_for_nifti_file
MedImage3D.formulate_qto_xyz
MedImage3D.check_sform_qform_similarity
MedImage3D.crop_mi
MedImage3D.calculate_qfac
MedImage3D.translate_mi
MedImage3D.formulate_nifti_image_struct
MedImage3D.formulate_spacing_scale_for_xyzt_space
MedImage3D.formulate_header_data_dict
MedImage3D.pad_mi
```
Here since we are only dealing with one module MedImage3D, we are mentioning all the functions directly. Make sure to add modules followed by your package name, if they are introduced.
A potential workaround if the Pkg is not in the registry
If your package is not yet in the registry yet, we can develop it on our current julia installation, but this would result in the documentation to be hosted locally. Make sure there are not package dependency errors within your Project.toml file in the root of the package. Remove unnecessary dependencies before beginning forward. Make sure a branch named "gh-pages" (example, replace this with the name of the branch you entered inside of your make.jl file in the docs) is present in this repository.
clone the main branch of medimage and change into that directory
run julia
enter pkg mode by hitting ]
run command develop ./
get out of pkg mode by hitting backspace
run command using MedImage3D
Voila, your package is now available within your julia environment.
Now in order to deploy your docs, within the same julia environment execute the following :
now continuting in the same julia repl
run command cd("docs")
enter package mode by hitting ]
run command activate . (with a dot)
rum command add DocumenterVitepress, Documenter
hit backspace to get out of pkg mode
run command include("make.jl")
enter shell mode by hitting ; (colon)
run command npm i
hit backspace to exit shell mode
finally run command DocumenterVitepress.dev_docs("build")
now finally open browser and navigate to the
https://localhost:5173/MedImage.jl/ (careful with the last / need to be added)
take a look at the locally deployed docs
TO EDIT AND ADD DOCUMENTATION
Edit or add markdown files within the src of docs, and update the make.jl file accordingly for the pages variable.
--docs/src/contributing.md
--docs/src/tutorials.md
Steps for troubleshooting
Remove the Manifest.toml file from the docs/ directory and try the process of execution again.
Update the make.jl file in the docs in case the repository name and github url gets changed.
Understanding about dependency resolution within Project.toml files.
In cases of Unsatisfied dependency tree issues , try the following :
Guide for handling unsatisfied requirement dependencies
Make sure to edit your Project.toml file for the package to remove any unneccessary dependencies if they show up in the unsatisfied dependency requirement error tree.
Fruit of work (a successful Pull Request)
Added Documentation Support for Medimage3D.jl
Migrating Documentation from Documenter to DocumenterVitepress
Subscribe to my newsletter
Read articles from Divyansh Goyal directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by

Divyansh Goyal
Divyansh Goyal
Microsoft Learn Student Ambassador Sentient Being | Julia Lang admirer