A Step-by-Step Guide to Decision Trees in Machine Learning
 Arbash Hussain
Arbash Hussain
Introduction
Welcome back, everyone! Apologies for the delay in continuing our ML algorithms series that started on January 1, 2024. But no worries, we are back on track and ready to speed things up! From now on, I’ll be uploading one machine learning algorithm-based blog every week.
So lets get started, In this blog post, we will build a Decision Tree model from scratch, explaining each and every step and later testing the model on Breast Cancer dataset. By the end, you’ll have a solid understanding of Decision Trees and how to implement them in code.
What is a Decision Tree?
A Decision Tree is a type of supervised learning algorithm used for both classification and regression tasks. It works by splitting the data into subsets based on the value of input features, making decisions at each node until reaching a final prediction at the leaf nodes. Lets understand this with the help of a hypothetical scenario.
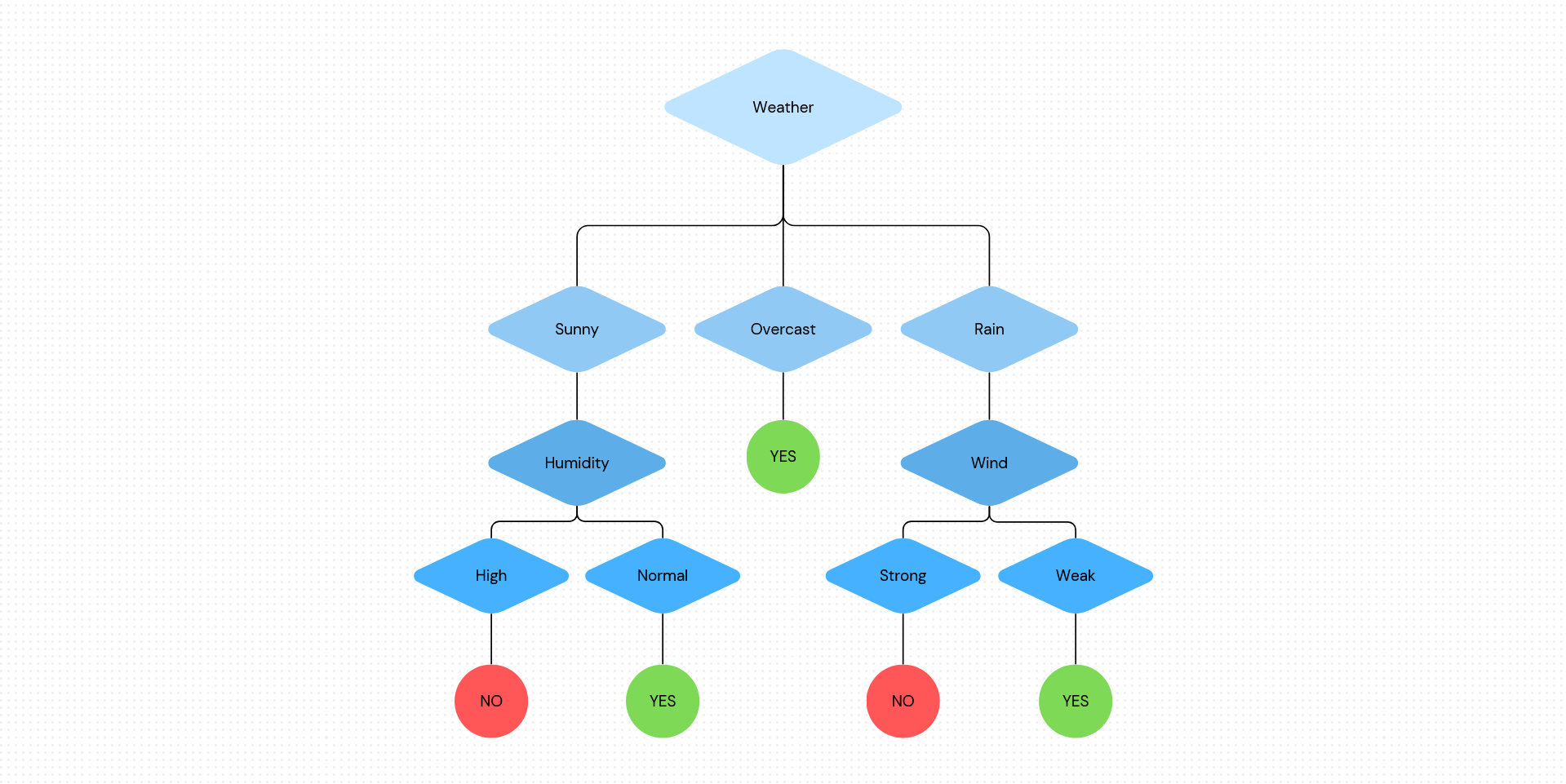
The following diagram illustrates the flow of decision tree for decision making with labels.
Rain = Yes
No Rain = No
How Splitting Happens
The way a Decision Tree decides how to split the data involves different techniques:
Gini Impurity (for Classification):
- Think of Gini Impurity as a measure of how mixed up the labels are in a group. If you randomly pick an item from a group, Gini Impurity tells you the chance of it being mislabeled. Lower Gini Impurity means the group is more pure, with mostly the same labels.
Information Gain (for Classification):
- Information Gain is like tidying up messy information. It uses entropy, which is a measure of chaos or randomness. By splitting the data based on a feature, we aim to make the subsets more organized and less random. Higher Information Gain means the data becomes more ordered after the split. We'll use this technique for our implementation.
Mean Squared Error (for Regression):
- Imagine you're trying to predict someone's weight. Mean Squared Error measures how far off your predictions are from the actual weights, squared (to make all differences positive). Lower MSE means your predictions are closer to the truth, minimizing the overall error.
Mean Absolute Error (for Regression):
- Mean Absolute Error is similar to MSE, but instead of squaring the differences, we just take their absolute values. This gives us a measure of how much, on average, our predictions differ from the actual values. Lower MAE means our predictions are more accurate, with smaller errors on average.
These techniques help the Decision Tree decide the best way to split the data at each step, ensuring that the final tree is as accurate and efficient as possible.
Key Concepts
Nodes and Leaves: Each decision point in the tree is called a node, and the final output points are called leaves.
Splitting: Dividing a node into two or more sub-nodes based on certain criteria.
Entropy and Information Gain: Metrics used to decide the best split. Entropy measures the randomness / impurity in a dataset, and information gain calculates the reduction in entropy after the dataset is split on an attribute, so more the information gain the better our split is.
Stopping Criteria:
Maximum no of layer of nodes our decision tree can grow.
Minimum no of samples a node must have if samples are less then don’t split the node.
Minimum entropy change for a split to take place.
Step-by-Step Implementation
Step 1: Import Necessary Libraries
We’ll use NumPy for numerical operations, Pandas for data manipulation, and some utilities from Scikit-learn for loading datasets and splitting data.
import numpy as np
from collections import Counter
from sklearn import datasets
from sklearn.model_selection import train_test_split
Step 2: Define the Node Class
The Node class represents each node in the tree.
class Node:
def __init__(self,feature=None,threshold=None,left=None,right=None,*,value=None):
self.feature = feature
self.threshold = threshold
self.left = left
self.right = right
self.value = value # value of node is Yes/No only incase of Leaf, else None
def is_leaf(self):
return self.value is not None # Returns true if the node is leaf else false
Each node stores information about the feature and threshold for splitting, pointers to left and right child nodes, and the value if it is a leaf node.
Step 3: Define the Decision Tree Class
class Decision_Tree:
def __init__(self, max_depth=10, min_sample_split=2, criteria='entropy', n_features=None):
self.max_depth = max_depth # Stopping Criteria
self.min_sample_split = min_sample_split # Stopping Criteria
self.criteria = criteria # Criteria type, entropy in our case.
self.n_features = n_features # No of features we'll be using for constructing the tree.
self.root = None
Training the Tree
The fit method trains the tree on the provided dataset.
def fit(self, X_train, y_train):
self.n_features = X_train.shape[1] if not self.n_features else min(X_train.shape[1], self.n_features)
# So that the no of features in the tree do not exceed the actual no of features we have in data.
n_samples = X_train.shape[0] # No of samples.
self.root = self.construct_tree(X_train, y_train, self.n_features, n_samples)
Here
X_train.shape[0]gives us no of samples/rows.X_train.shape[1]gives us no of features/columns.
Constructing the Tree
The construct_tree method recursively builds the tree by splitting the data.
def construct_tree(self, X, y, n_features, n_samples, depth=0):
# No of labels there are in a specific feature,
# if 1 then no need to split.
# For eg. in case of Wind, we can go to 2 labels (strong and weak) so we split.
labels = len(np.unique(y))
# Check the stopping criteria
# if met create a leaf node, based on label with max frequence
if depth >= self.max_depth or labels == 1 or n_samples < self.min_sample_split:
leaf_value = self.most_common_label(y)
return Node(value=leaf_value)
# Find the best split
feat_indexs = np.random.choice(n_features, self.n_features, replace=False)
best_threshold, best_feature = self.best_split(X, y, feat_indexs)
# Create child nodes (Recursively Create Tree)
left_indxs, right_indxs = self.split(X[:, best_feature], best_threshold)
left = self.construct_tree(X[left_indxs, :], y[left_indxs], n_samples, depth + 1)
right = self.construct_tree(X[right_indxs, :], y[right_indxs], n_samples, depth + 1)
return Node(best_feature, best_threshold, left, right)
def most_common_label(self,y):
c = Counter(y)
return c.most_common(1)[0][0]
Here we first check whether the stopping criteria is met.
If the stopping criteria is met, we'll create a leaf node.
If the stopping criteria is not met, we'll find the best split.
Create left and right child based on the best split.
Splitting the Data
The split method divides the data based on the threshold.
def split(self,X_col,threshold):
# Left Split, val <= threshold
left_idxs = np.argwhere(X_col<=threshold).flatten()
# Right Split, val> threshold
right_idxs = np.argwhere(X_col>threshold).flatten()
# np.argwhere returns the indices in a list of lists, so we flatten the result.
return left_idxs, right_idxs
left_idxsare the indices of X_col having val less than or equal to threshold.right_idxsare the indices of X_col having val more than threshold.
Finding the Best Split
The best_split method iterates over all features and thresholds to find the best split based on information gain.
def best_split(self,X,y,feat_indexs):
best_gain = -1
split_index = None
split_threshold = None
for feat_index in feat_indexs:
X_col = X[:, feat_index] # Values of feature X_col.
thresholds = np.unique(X_col) # Selecting all the unique values of X_col as thresholds.
for thr in thresholds:
# For each threshold in thresholds.
# Find the threshold with maximum Information Gain
gain = self.calculate_gain(X_col,y,thr)
if gain > best_gain:
best_gain = gain
split_index = feat_index
split_threshold = thr
return split_threshold, split_index
Initially our
best_gain,split_index,split_thresholdare set to -1, None, None.For a
X_colwe get all its unique vals in a list, and find which amongst them will be the best threshold for the split.
Calculating Entropy and Information Gain
In this blog we use Information gain for splitting the nodes in decision tree. Information Gain is calculated using the formula:
Information Gain = Entropy(Parent) - Weighted Average * Entropy(Children)
Where:
Entropy(Parent)is the entropy of the parent node.Weighted Averageis the average of entropy of the children, weighted by their number of instances.Entropy(Children)is the entropy of each child node.
Entropy is calculated using the formula:
Entropy = - ∑ [ p(x) * log(p(x)) ]
Where:
p(x)is the probability of occurrence of an event, i.e. number of times classxhas occurred divided by total no of samples.p(x) = count(x) / n
def calculate_entropy(self, y):
hist = np.bincount(y) # returns a frequency list of elements, from 0 to max(y).
ps = hist / len(y) # [p(x1), p(x2), p(x3),..., p(xN)]
return -np.sum([p * np.log(p) for p in ps if p > 0]) # Only consider non-zero probabilities
def calculate_gain(self, X_col, y, threshold):
entropy_parent = self.calculate_entropy(y)
# Create children
left_idxs, right_idxs = self.split(X_col, threshold)
if len(left_idxs) == 0 or len(right_idxs) == 0:
return 0
# Calculate the weighted average entropy of the children.
n = len(y)
# No of samples in left and right.
n_l, n_r = len(left_idxs), len(right_idxs)
# Left entropy and right entropy.
e_l, e_r = self.calculate_entropy(y[left_idxs]), self.calculate_entropy(y[right_idxs])
# No of samples in left/total samples times left entropy + No of samples in right/total samples times right entropy.
weighted_avg_entropy_children = (n_l / n) * e_l + (n_r / n) * e_r
# Calculate Information Gain
info_gain = entropy_parent - weighted_avg_entropy_children
return info_gain
Making Predictions
The predict method traverses the tree to make predictions on new data.
def predict(self, X):
return np.array([self.traverse_tree(x, self.root) for x in X])
def traverse_tree(self, X, node):
if node.is_leaf():
return node.value
if X[node.feature] <= node.threshold: # Recursively travel
return self.traverse_tree(X, node.left) # val <= threshold, Goto Left child.
return self.traverse_tree(X, node.right) # val > threshold, Goto Right child.
We send a data
X, to predict function.The data travels the tree until it reaches the predicted leaf.
Step 4: Testing the Model
Finally, we test our Decision Tree model on the breast cancer dataset.
def mse(y1, y2):
return np.mean((y2 - y1)**2)
def accuracy(y1,y2):
return np.sum(y1==y2)/len(y1)
if __name__ == "__main__":
model = Decision_Tree()
data = datasets.load_breast_cancer()
X, y = data.data, data.target
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
model.fit(X_train, y_train)
preds = model.predict(X_test)
error = mse(y_test, preds)
print('Error:', error)

Performance Evaluation on Breast Cancer Dataset
Testing Accuracy - 0.88 or 88%
Mean Squared Error - 0.114
Points to Note: Common Misconceptions
Misconception 1: Decision Trees Are Always Prone to Overfitting
While Decision Trees can be prone to overfitting, especially with deep trees that capture noise in the training data, this is not always the case. Proper pruning techniques and parameter tuning can significantly mitigate overfitting.
Explanation: The misconception arises because Decision Trees are flexible models that can adapt closely to the training data. However, by setting constraints like maximum depth and minimum samples per leaf, or by using ensemble methods like Random Forests, we can control overfitting effectively.
Misconception 2: Decision Trees Are Always Better with More Features
Adding more features to a Decision Tree does not always improve its performance. Irrelevant or redundant features can confuse the model and lead to poorer performance.
Explanation: Including too many features, especially those that do not contribute meaningful information, can lead to a more complex tree with unnecessary splits. Feature selection techniques or regularization methods can help identify the most relevant features for building a robust model.
When to Apply Decision Trees: Key Points to Consider
Non-linearity: Decision Trees do not require the relationship between input and output variables to be linear. They can very well handle non-linear relationships and interactions between features.
Handling of Mixed Data Types: Decision Trees can handle both numerical and categorical data, making them versatile for different types of datasets.
Handling Missing Values: Decision Trees can manage missing values in the data. During the splitting process, they can work without requiring imputation.
Robustness to Outliers: Decision Trees are relatively robust to outliers, as splits are based on feature thresholds that can separate outliers from the majority of the data.
Small to Medium-Sized Data: Decision Trees work well with small to medium-sized datasets. However, for very large datasets, ensemble methods like Random Forests or Gradient Boosting Trees might be more efficient, which we'll discuss soon.
Subscribe to my newsletter
Read articles from Arbash Hussain directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by

Arbash Hussain
Arbash Hussain
I'm a Computer Science Engineer with a passion for data science and AI. My interest for computer science has motivated me to work with various tech stacks like Flutter, Next.js, React.js, Pygame and Unity. For data science projects, I've used tools like MLflow, AWS, Tableau, SQL, and MongoDB, and I've worked with Flask and Django to build data-driven applications. I'm always eager to learn and stay updated with the latest in the field. I'm looking forward to connecting with like-minded professionals and finding opportunities to make an impact through data and AI.