PacBio SMRT Sequencing for Human Telomere Sequence
 kikogarcia
kikogarciaIntroduction
Telomeres are specialized nucleoprotein structures at the ends of linear chromosomes, and the ends of human chromosomes are covered by telomeres. Telomeres contain approximately 5-15 kb of TTAGGG-rich double-stranded repetitive sequences and a shelterin protein complex that protects the chromosome ends, with G-rich single-stranded overhangs at the 3' end, and are essential for maintaining the stability and intact replication of the human genome.
Telomeres are primarily dependent on telomerase for their synthesis and are not constant in length. With the exception of germ cells and stem cells, somatic cells have a progressive shortening of telomeres (~50-200 bp) with each cell division due to a deficiency in telomerase activity. As we age, telomeres shorten slowly from early life to adulthood, and when telomeres become very short, they become dysfunctional, inducing a DNA damage response, leading to cellular senescence and a range of diseases. Therefore, the progressive shortening of telomere length in normal human cells is a promising biomarker for age-related diseases.
Telomeres and filaments have long puzzled genome scientists. In the early days of genome sequencing, many researchers took for granted that assembling these highly repetitive regions was essentially impossible. Multiple telomere detection technologies failed to achieve high-throughput detection of telomeres at single-base resolution or to accurately quantify telomere length. As a result, there is an urgent need for a highly sensitive method for the timely longitudinal detection of individual telomere wear in both experimental and clinical studies, and the emergence of Pacific Biosciences (PacBio) Single Molecule Real-Time (SMRT) sequencing technology represents a transformative leap forward in this field, opening up new possibilities for comprehensive and accurate telomere sequencing.
Our leading-edge PacBio SMRT sequencing technology platform allows you to accurately measure the intractable regions of the human genome—telomere sequences.
Challenges to Sequencing Human Telomeres
Historically, techniques such as terminal restriction fragment (TRF) analysis, real-time quantitative polymerase chain reaction (qPCR), high-throughput single telomere length analysis (HT-STELA), and high-throughput fluorescence in situ hybridization (HT-QFISH) have been used to measure telomere length. However, these methods only provide average or relative telomere length and lack the precise single-base resolution required for a nuanced understanding of telomere biology. More importantly, they do not take into account telomere variant sequences (TVS) that can significantly affect human aging and disease.
Long read-length sequencing platforms have the potential to overcome these limitations, enabling direct sequencing of telomeric genomic DNA fragments and determination of absolute telomere lengths at the ends of individual chromosomes. However, such sequencing methods require whole genome sequencing, making high-throughput analysis impractical. They also fail to maintain the integrity of the terminal 3' G-rich prominence, further complicating the sequencing process.
Telomere Genome Analysis with PacBio SMRT Sequencing
PacBio SMRT sequencing technology offers an elegant solution to these challenges, opening up new frontiers in human telomere research. SMRT sequencing is capable of generating long reads of tens of thousands of base pairs (known as HiFi reads) in a single extension, greatly reducing the alignment challenges associated with highly repetitive regions such as telomeres.
By utilizing a novel workflow called "Telobaits," researchers have been able to capture telomeres from cell lines and clinical patient samples. The Telobait workflow employs six newly designed telomeric DNA sequences that contain repeats complementary to the protruding ends of telomeric 3' single strands. The process involves capturing chromosome ends with Telobait and purifying full-length telomeric genomic DNA fragments with streptavidin-coupled magnetic beads. The PacBio HiFi sequencing platform enables high-throughput telomere length measurements at single-base resolution.
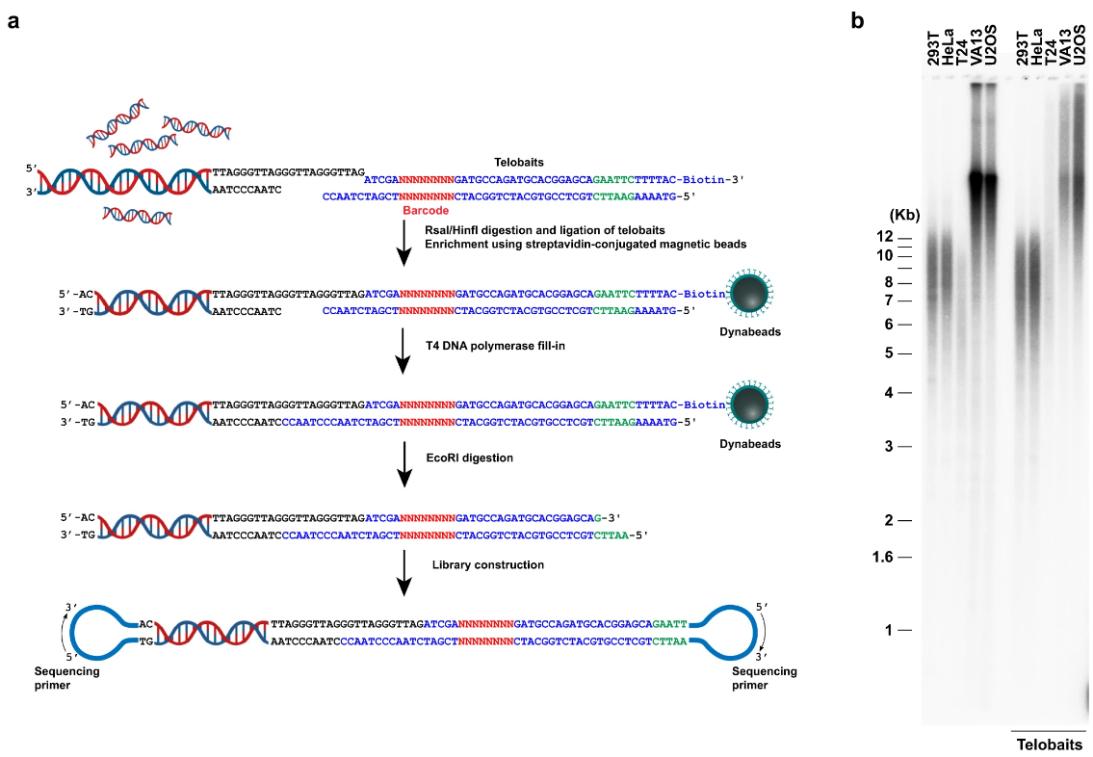
Enrichment of telomere-containing genomic DNA fragments using telobaits for PacBio HiFi sequencing. (Tham C Y et al., 2023)
Advantages of PacBio SMRT Sequencing for Telomere Analysis
Provides high-throughput, accurate telomere length measurement at single-base resolution.
Allows for a more comprehensive examination of telomere variant sequences (TVS), a key determinant of the aging process and the pathophysiology of many diseases.
The high-throughput telomere length measurement capability of SMRT sequencing allows for large-scale studies.
The long read-length sequencing method used in SMRT sequencing accurately analyzes the G-strand at the end of chromosomes.
References
- Tham, Cheng-Yong, et al. "High-throughput telomere length measurement at nucleotide resolution using the PacBio high fidelity sequencing platform." Nature Communications 14.1 (2023): 281.
Subscribe to my newsletter
Read articles from kikogarcia directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
kikogarcia
kikogarcia
website: https://www.cd-genomics.com/ CD Genomics is a leading global life sciences company, and we remain committed to providing the research community with high-quality long-read sequencing services, from Oxford Nanopore to PacBio SMRT sequencing.