Advancing RAG with unstructured.io
 Praghadeesh T K S
Praghadeesh T K S
Hello All, This is Praghadeesh back to writing blogs after a while (I lost my previously hosted Ghost Instance with no backups and had to start from scratch 😕). In this blog, let's explore a bit more on RAG by trying to work on some complex PDFs leveraging the capabilities of unstructured.io and Langchain's MultiVectorRetriever this time.
What is unstructured.io?
Unstructured.io is an open source project that provides tools with capabilities to work on diverse source of documents such as PDF, HTML and so on and helps us to streamline the data processing workflow for LLMs. It's more of an ETL tool for Gen AI use cases. It comes in three different offerings
Serverless API
Azure/AWS Marketplace offering
Self hostable solution
What is RAG and why to use unstructured.io with RAG?
If the title of the blog interested you and you are already here reading the blog, you might probably know what RAG is all about. In oversimplified terms, it's just the art of injecting context to the LLMs where the goal is to help them answer questions that is beyond the training data of LLM. I believe this might be a perfect analogy, It's like an Open Book Exam, where you try to find the relavant content from the book and make sense out of it.
Cool, but how unstructured helps here? The process of RAG becomes complex when we try to deal with diverse contents such as Tables, Images, Vector Diagrams, Formulae and so on. Unstructured.io will help us to work with some of these data and make our job a bit easier, the scope of the blog is limited to handing the data in tabular format in complex PDFs.
Working with Complex PDFs
Complex PDFs may involve Financial Reports, Scientific Research papers, Technical Reference Document, Engineering Datasheets and so on. In this blog, let's try dealing with a datasheet for an Electrical component called LM-317 a linear voltage regulator.
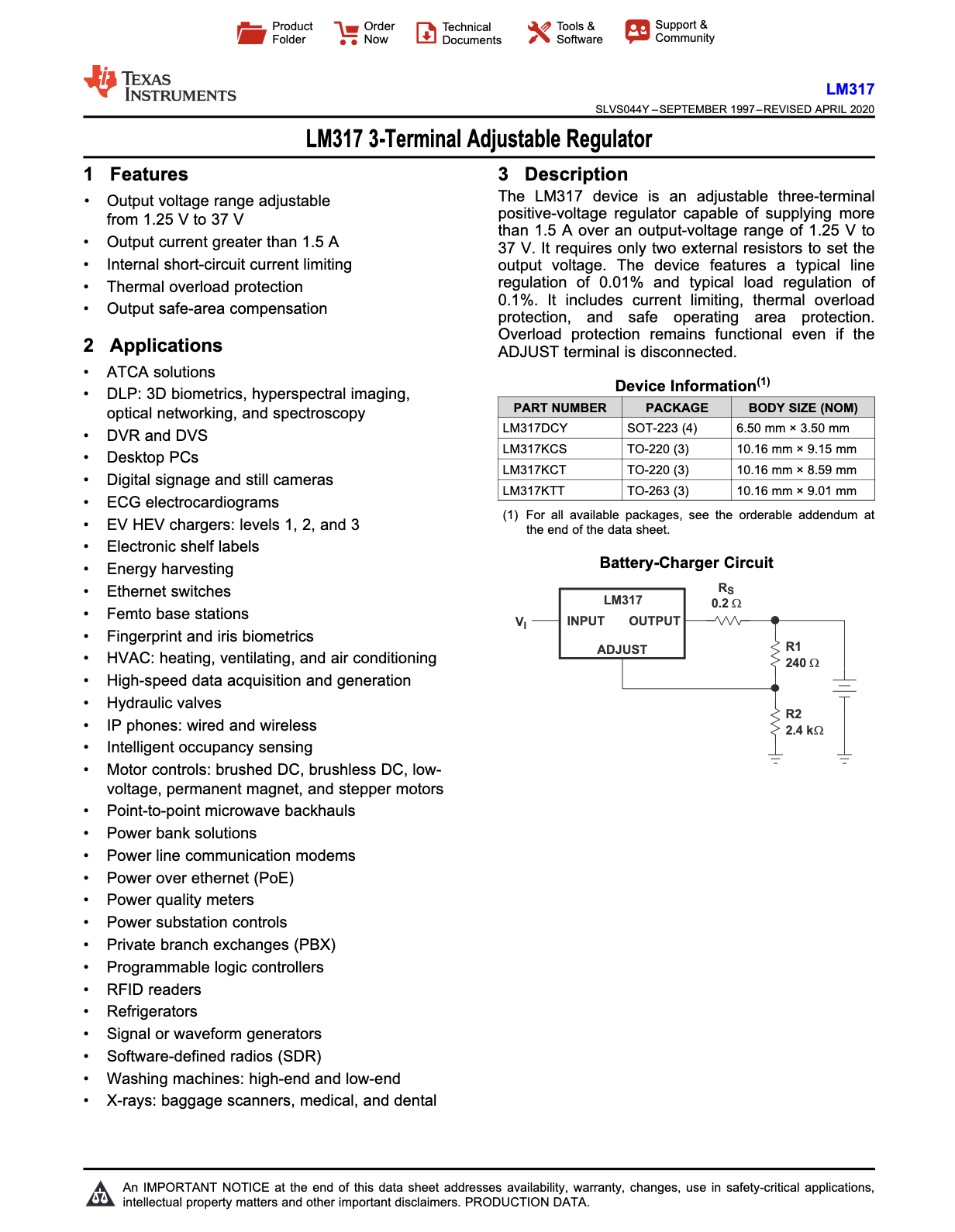
The above is an example of how the content of the datasheet looks like, it has multiple pages with such tables and vector diagrams where extracting data without loosing quality might not be possible with traditional RAG.
Semi Structured RAG with Multi Vector Retreiver
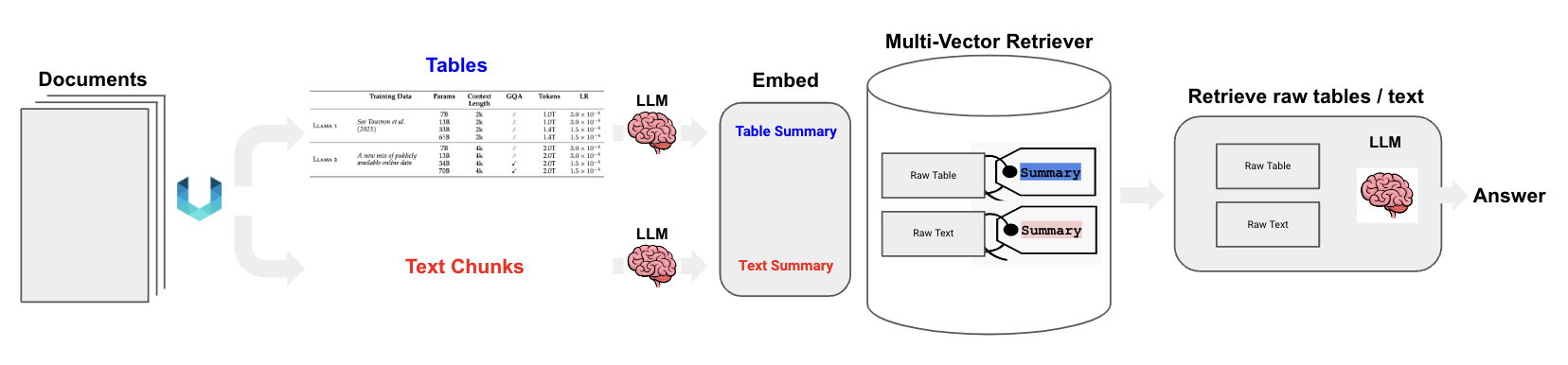
The idea here is to extract text and table chunks separately as shown above using unstructured
Create a summarization chain and generate summary for texts and tables
Ingest the text and table summary with corresponding embeddings into the vector store
Ingest the Raw chunks into the docstore or memorystore
Query against the summary embeddings, retrieve the corresponding chunks from docstore associated with summary in vectorstore and pass the chunk to the LLM to make sense out of it
Partitioning the PDF document using unstructured
unstruct_client = UnstructuredClient(
api_key_auth=os.getenv("UNSTRUCTURED_API_AUTH_KEY")
)
filename = "lm317.pdf"
with open(filename, "rb") as f:
files=shared.Files(
content=f.read(),
file_name=filename,
)
req = shared.PartitionParameters(
files=files,
strategy="hi_res",
hi_res_model_name="yolox",
skip_infer_table_types=[],
pdf_infer_table_structure=True
)
try:
resp = unstruct_client.general.partition(req)
pdf_elements = dict_to_elements(resp.elements)
except SDKError as e:
print(e)
In the above part of code, the partitioning of PDF is executed. Unstructured simplifies the preprocessing of structured and unstructured documents for downstream tasks, irrespective of what type of file content is provided as source. When partioned the result is a list of Element objects.
The below is an example of how the Partition output looks like, the Elements can be of type Title, NarrativeText, Image, Table, ListItem, Header, Footer and so on.
{
"type":"Title",
"element_id":"d8ecdee23702fdb35f98390141100d13",
"text":"from 1.25 V to 37 V",
"metadata":{
"filetype":"application/pdf",
"languages":[
"eng"
],
"page_number":1,
"filename":"lm317.pdf"
}
},
{
"type":"ListItem",
"element_id":"eb105b9f3e577473acac7ba394cea3c7",
"text":"Output current greater than 1.5 A • • Thermal overload protection • Output safe-area compensation",
"metadata":{
"filetype":"application/pdf",
"languages":[
"eng"
],
"page_number":1,
"parent_id":"d8ecdee23702fdb35f98390141100d13",
"filename":"lm317.pdf"
}
},
Chunking the elements obtained after partitioning
The partitions created are then chunked using the chunking strategy - chunk_by_title.
by_title chunking strategy preserves section boundaries and optionally page boundaries as well. “Preserving” here means that a single chunk will never contain text that occurred in two different sections. When a new section starts, the existing chunk is closed and a new one started, even if the next element would fit in the prior chunk.The chunks are categrozied as table chunks and text chunks respectively and a summary chain is created using the Google Gemini Pro model, which will help us creating a list of table summaries and text summaries.
chunks = chunk_by_title(pdf_elements,max_characters=4000,new_after_n_chars=3800, combine_text_under_n_chars=2000)
class Element(BaseModel):
type: str
text: Any
# Categorize by type
categorized_elements = []
for element in chunks:
if "unstructured.documents.elements.Table" in str(type(element)):
categorized_elements.append(Element(type="table", text=str(element)))
elif "unstructured.documents.elements.CompositeElement" in str(type(element)):
categorized_elements.append(Element(type="text", text=str(element)))
# Tables
table_elements = [e for e in categorized_elements if e.type == "table"]
print(len(table_elements))
# Text
text_elements = [e for e in categorized_elements if e.type == "text"]
print(len(text_elements))
# Prompt
prompt_text = """You are an assistant tasked with summarizing tables and text. \
Give a concise summary of the table or text. Table or text chunk: {element} """
prompt = ChatPromptTemplate.from_template(prompt_text)
# Summary chain
model = ChatGoogleGenerativeAI(model="gemini-pro")
summarize_chain = {"element": lambda x: x} | prompt | model | StrOutputParser()
# Apply to tables
tables = [i.text for i in table_elements]
table_summaries = summarize_chain.batch(tables, {"max_concurrency": 5})
# Apply to texts
texts = [i.text for i in text_elements]
text_summaries = summarize_chain.batch(texts, {"max_concurrency": 1})
Adding the Summaries and Documents to Vector Store and Doc Store
The summaries are added to the vector store (ChromaDB in this case) and the raw chunks are added to the docstore both mapped with a uid.
# The vectorstore to use to index the child chunks
vectorstore = Chroma(collection_name="summaries", embedding_function=FastEmbedEmbeddings())
# The storage layer for the parent documents
store = InMemoryStore()
id_key = "doc_id"
# The retriever (empty to start)
retriever = MultiVectorRetriever(
vectorstore=vectorstore,
docstore=store,
id_key=id_key,
)
# Add texts
doc_ids = [str(uuid.uuid4()) for _ in texts]
summary_texts = [
Document(page_content=s, metadata={id_key: doc_ids[i]})
for i, s in enumerate(text_summaries)
]
retriever.vectorstore.add_documents(summary_texts)
retriever.docstore.mset(list(zip(doc_ids, texts)))
# Add tables
table_ids = [str(uuid.uuid4()) for _ in tables]
summary_tables = [
Document(page_content=s, metadata={id_key: table_ids[i]})
for i, s in enumerate(table_summaries)
]
retriever.vectorstore.add_documents(summary_tables)
retriever.docstore.mset(list(zip(table_ids, tables)))
Creating the answer chain
As a final process, the RAG chain is created and the query is passed as an input to the RAG chain.
# Prompt template
template = """Answer the question based only on the following context, which can include text and tables:
{context}
Question: {question}
"""
prompt = ChatPromptTemplate.from_template(template)
# LLM
model = ChatGoogleGenerativeAI(model="gemini-pro")
# RAG pipeline
chain = (
{"context": retriever, "question": RunnablePassthrough()}
| prompt
| model
| StrOutputParser()
)
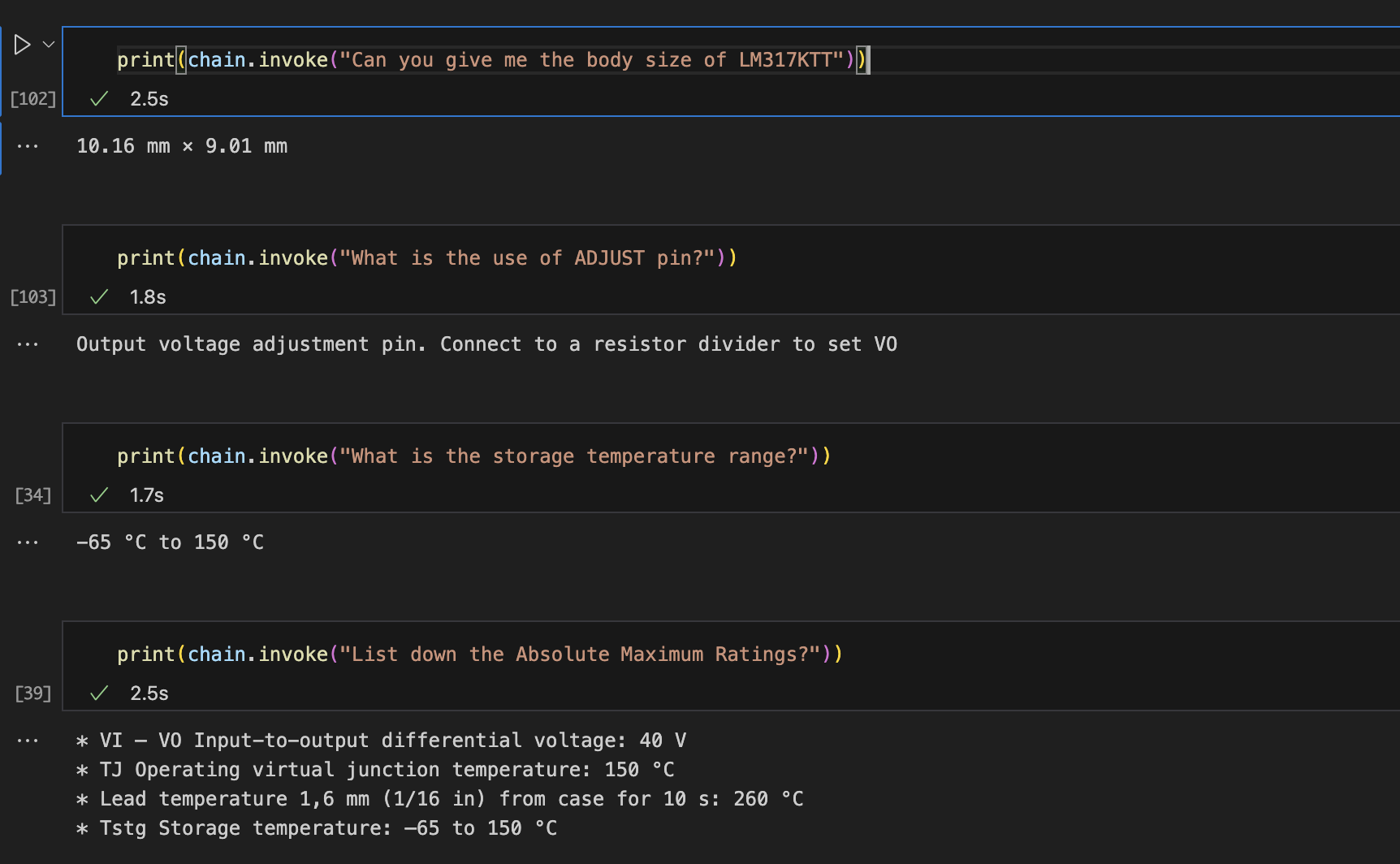
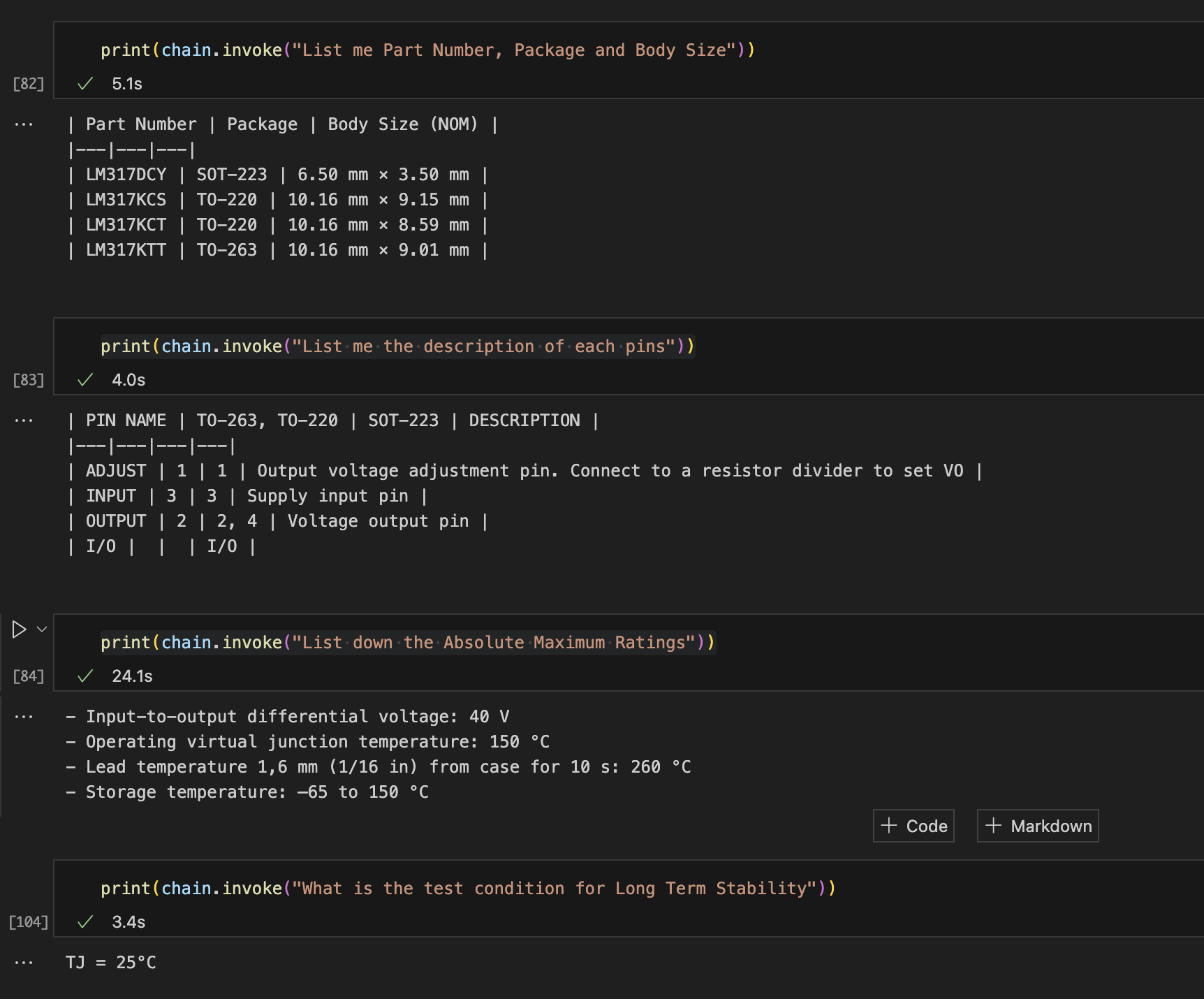
As we can see above, the LLM Chain is able to provide us with accurate results from the tables present in the datasheet of LM317 Linear Voltage Regulator.
References
1. Semi Structured RAG Cookbook
2. Unstructured IO Documentation
Subscribe to my newsletter
Read articles from Praghadeesh T K S directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
