Guide to Principal Component Analysis in Data Science
 Adeniran Emmanuel
Adeniran Emmanuel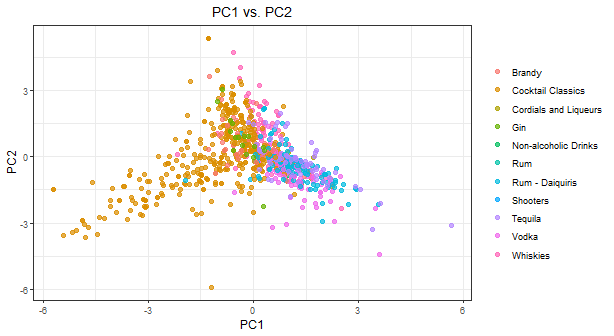
Origin of PCA
Approach of PCA
How to do PCA
When is PCA used
What PCA is and is not
Advantages and Disadvantages of PCA
Metric of Evaluation
After PCA, what Next?
Case Study of the Cocktail recipe Dataset
Origin of PCA
PCA was first developed by Karl Pearson in 1901 and later developed by Hotelling in 1933.Pearson was a statistician and what he was working on when he developed PCA isn't entirely documented but we can suggest two things that could have prompted the development of the mathematical technique which are:
Biometry(The application of Statistical methods to Biology)
Factor Analysis
PCA applies an Orthogonal linear transformation to reframe the original correlated data in a new coordinate framework called Principal Components which are linearly uncorrelated. The largest variance(at least 80%) is usually explained in the First Component i.e. PC1 and the least component has the least Variance
Approach of PCA
There are two main approach of the PCA which are:
Eigenvalue decomposition of the data covariance matrix
Singular-Value Decomposition of the centered Data matrix
PCA is commonly used when many of the variables are highly correlated with each other and it's desirable to reduce their number to an independent set, the idea of PCA is to reduce the number of variables of a dataset while preserving as much information as possible
How to do PCA
This steps are done in the backend of the tool used to create the Principal Components
Standardize the range of continuous initial variables
Compute the covariance matrix to identify correlation
Compute the Eigenvalues and Eigenvectors of the covariance matrix to identify the Principal Components
Create a feature vector to decide which Principal Components to keep
Replot the data along the PC axes
When is PCA used?
PCA is used when we wish to reduce the dimensionality of a large dataset to a small and explains important information in the data
What PCA is and isn't
PCA is:
Is a Mathematical Technique and an Unsupervised Learning method(i.e. we use unlabeled data to process our data)
Is a Dimensionality Reduction Technique
PCA is not:
Not a data Cleaning method
Not a data Visualization technique
Not a data Transformation Technique
Not a feature selection method
Not a Model or algorithm
Not a perfect solution to all cases
Advantages and Disadvantages of PCA
Advantages
Removes Noise from Data
Removes Multicollinearity
Reduces Model Parameter
Improves Model Performance
Reduces Computational Cost
Disadvantages
No Feature Interpretability
Only offers Linear Dimensionality Reduction
Affected by Outliers
Loss of Information
High Run-time
Metrics of Evaluation
There isn't a single Evaluation Metric that is generally accepted for PCA, as it measures the proportion of the total variance in the data that is captured by the Principal Component.
Explained Variance
Scree Plot
Application-Specific Metric
After PCA, What Next?
After PCA has been done on a large dataset, the components can be used for the following
Visualization
Scatter Plot
Parallel Coordinates
Machine Learning - If you plan to use the data for ML task, PCA may be a good preprocessing task as using the transformed data can potentially improve model performance
Anomaly Detection - PCA can be used to identify data points that are significantly different from the lower-dimensional representation captured by the data
Case Study
Boston Cocktail Recipe Dataset
# Loading the Necessary Libraries
library(tidyverse) # A Powerful package used for Data Antasks
library(tidymodels) # A Powerful package used for Building Models
library(janitor)
# Loading the Dataset
boston <- read_csv("Machine Learning Dataset/boston_cocktails.csv")
glimpse(boston)
The glimpse() function helps us to understand the structure of our data, we can also use the skim(), skim_without_chart() function from the "skimr" package and many more
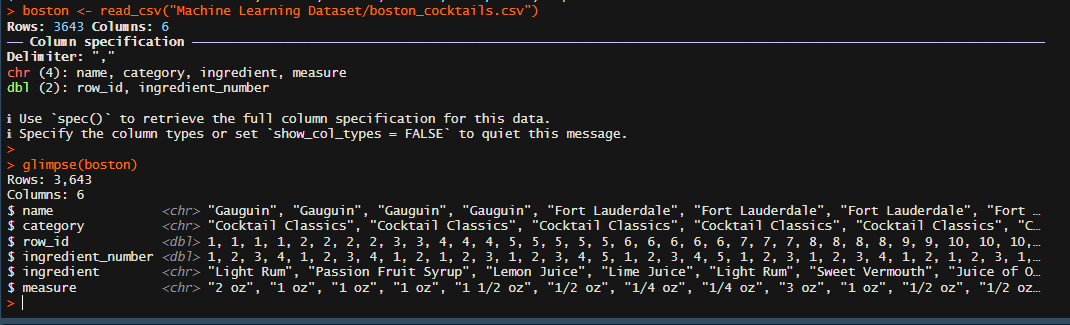
We have 4 character variables and 2 double variables, we still need to clean the data and put it in a format that we can run our Unsupervised learning method on
boston_parsed <-
boston |>
mutate(
ingredient = str_to_lower(ingredient),
ingredient = str_replace_all(ingredient, "-", " "),
ingredient = str_remove(ingredient, "liqueur|(if desired)"),
ingredient = case_when(
str_detect(ingredient, "bitters") ~ "bitters",
str_detect(ingredient, "orange") ~ "orange juice",
str_detect(ingredient, "lemon") ~ "lemon juice",
str_detect(ingredient, "lime") ~ "lime juice",
str_detect(ingredient, "grapefruit") ~ "grapefruit juice",
TRUE ~ ingredient
),
measure = case_when(
str_detect(ingredient, "bitters") ~ str_replace(measure, "oz$", "dash"),
TRUE ~ measure
),
measure = str_replace(measure, " ?1/2",".5"),
measure = str_replace(measure, " ?3/4", ".75"),
measure = str_replace(measure, " ?1/4", ".25"),
measure_number = parse_number(measure),
measure_number = ifelse(str_detect(measure, "dash$"),measure_number / 50, measure_number)
) |>
add_count(ingredient) |>
filter(n > 15) |>
select(-n) |>
distinct(row_id, ingredient, .keep_all = TRUE)
boston_parsed
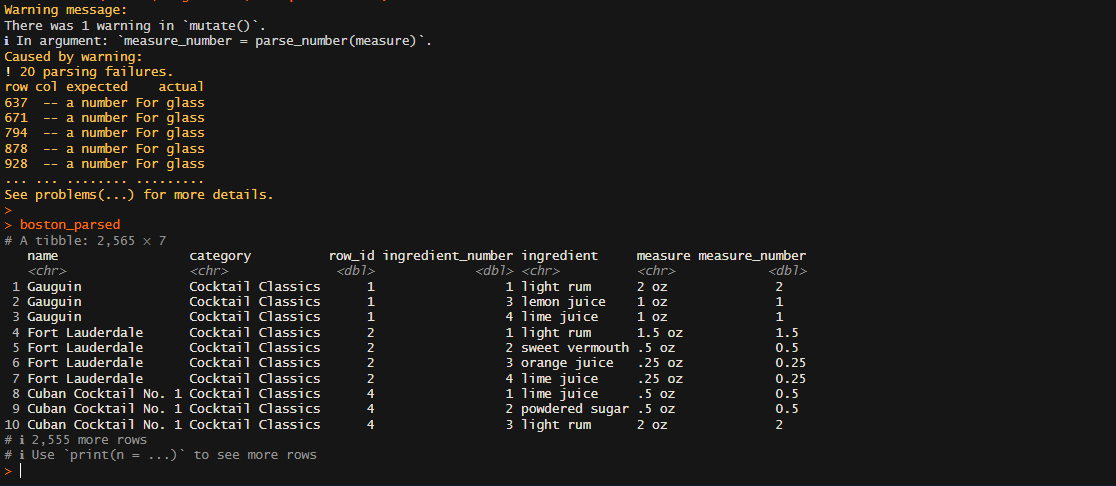
boston_df <-
boston_parsed |>
select(-row_id, -ingredient_number, -measure) |>
pivot_wider(names_from = ingredient,values_from = measure_number,
values_fill = 0) |>
clean_names() |>
na.omit()
boston_df
pivot_wider() is used to put our data in a wider format and the clean_names() is a function from the lubridate package
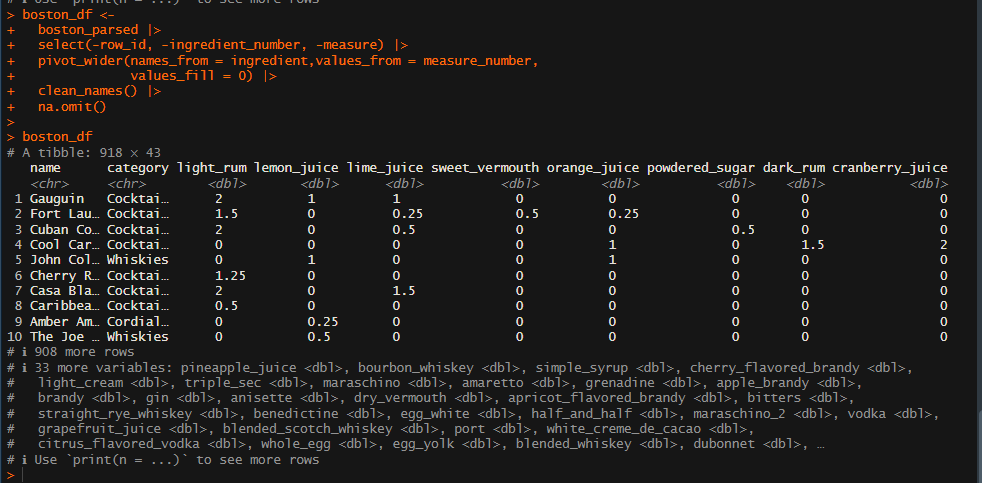
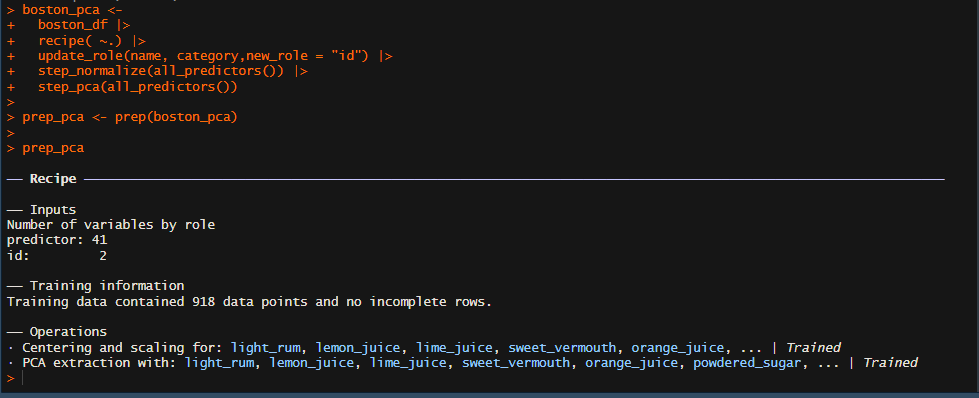
boston_pca <-
boston_df |>
recipe( ~.) |>
update_role(name, category,new_role = "id") |>
step_normalize(all_predictors()) |>
step_pca(all_predictors())
prep_pca <- prep(boston_pca)
prep_pca
The recipe() function is from the recipe package and offers numerous data preprocessing functions and steps we used the step_normalize() to center and scale our data and use the the update_role() to tell our algorithm that the columns selected are nor required in the learning and should be used only for "id" which can be seen in the image below
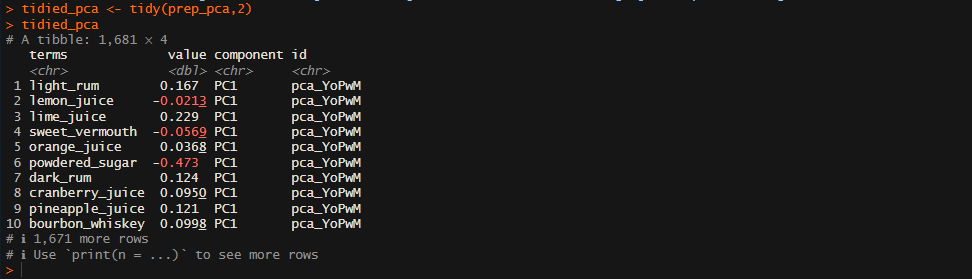
tidied_pca <- tidy(prep_pca,2)
tidied_pca
The tidy() function from the tidyr package can be used to extract our Principal Components
tidied_pca |>
filter(component %in% paste0("PC", 1:5)) |>
mutate(component = fct_inorder(component)) |>
ggplot(aes(value,terms, fill = terms)) +
geom_col(show.legend = FALSE) +
facet_wrap(vars(component), nrow = 1) +
labs(Y = NULL)
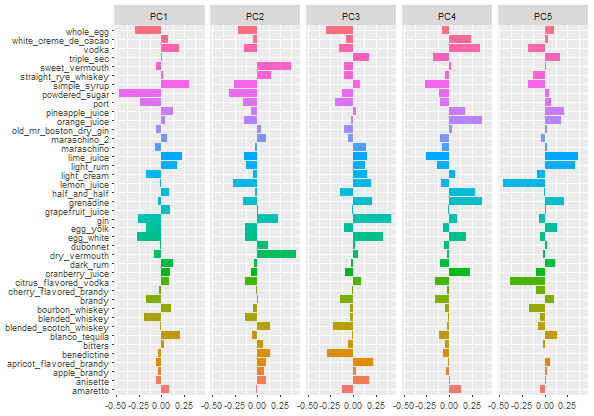
The final chart is showing the first 5 Principal Components and as stated earlier, the first 2 Principal Components i.e. PC1 and PC2 has more variance and explains more than 90% of the data dimensions
Subscribe to my newsletter
Read articles from Adeniran Emmanuel directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
