Using fsspec To Define OneLake Filesystem in Fabric
 Sandeep Pawar
Sandeep PawarI mentioned on X the other day that, like other filesystem backends such as S3 and GCS, you can use fsspec to define the OneLake filesystem too. In this blog, I will explain how to define it and why it's important to know about it.
First, I want to thank Pradeep Srikakolapu (Sr PM, Fabric CAT) for sharing this with me.
fsspec is a Python library that provides a unified interface for interacting with various filesystems, allowing users to use a consistent API for reading and writing data. For example, AWS has the S3 file system, Google Cloud has GCS, and Azure Blob storage has the abfs file system. By using fsspec, developers can keep their code consistent while simply changing the file system used. We will see an example below.
As you may already know, when you save any data to OneLake (files or tables), it has an abfs path. Users can use the abfs path to read and write data to any lakehouse. For example, a notebook could be in workspace A, but using the abfs path, you can read or write data to a lakehouse in workspace B without mounting the lakehouse or setting a default lakehouse. I have written about mounting and default lakehouse before.
You can use abfs path for reading data in spark as well as pandas & polars.
path = "abfss://<workspace_id>@onelake.dfs.fabric.microsoft.com/<lakehouse_id>/Files/*.parquet"
#spark
spark.read.format("parquet").load(path)
spark.sql("select * from <abfs_table_path>")
#pandas
pd.read_parquet(path)
#polars
pl.scan_parquet(path)
#dask
dd.read_parquet(path)
It's easy to create an abfs path if you know the workspace, lakehouse, schema and the relative path of the file/table you need to read/write. The advantage here is that you can programmatically define/parameterize the path which helps with branching and development.
What has fsspec to do with this? While spark, pandas, polars, Dask can read directly from an abfs path, not all Python libraries can. For example, if you try to use pyarrow or duckdb, it will fail. pyarrow doesn't recognize abfs filesystem:
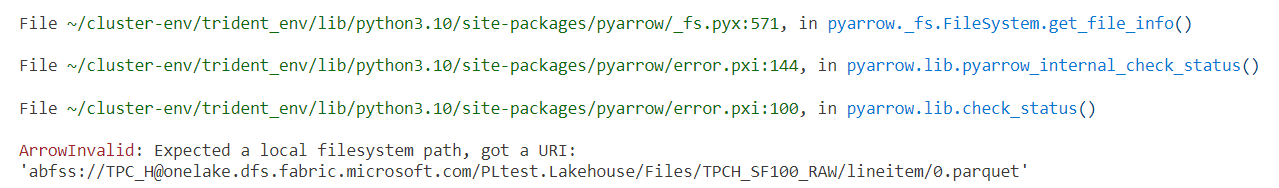
This is where we can define our custom filesystem and pass it to pyarrow as below:
- First, I create the abfss path programmatically. If you already have the abfs path, you can skip it.
%pip install duckdb --q
import sempy.fabric as fabric
import fsspec
import pyarrow.parquet as pq
import duckdb
def get_lakehouse_abfs(lakehouse_name, workspace_name, relative_path=None):
path = mssparkutils.lakehouse.get(name=lakehouse_name, workspaceId = fabric.resolve_workspace_id(workspace_name))['properties']['abfsPath']
if relative_path:
path = path + relative_path
return path
#Create the abfss path programmatically
path = get_lakehouse_abfs("product_sales_gold", "Product Sales", relative_path="/Files/covid-19_data.parquet")
Define the filesystem using
fsspec:filesystem_code = "abfss" storage_options = { "account_name": "onelake", "account_host": "onelake.dfs.fabric.microsoft.com" } onelake_fs = fsspec.filesystem(filesystem_code, **storage_options)Pass the filesystem to the library:
#pass the file system to pyarrow pq_table = pq.read_table(path, filesystem=onelake_fs)
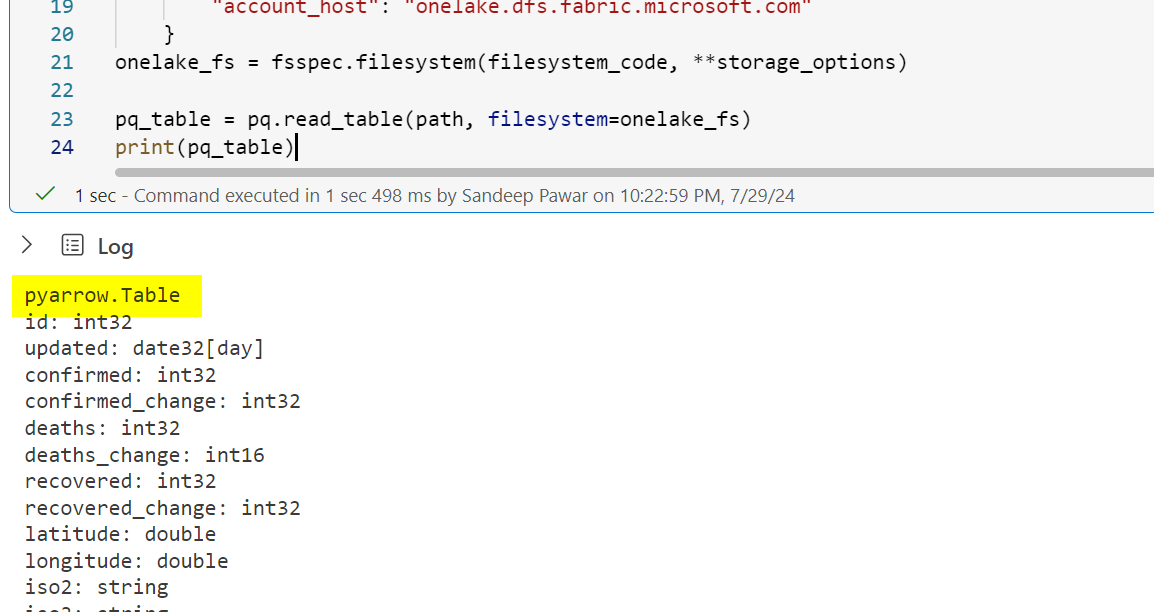
pyarrow is great for checking parquet schema and metadata, like I did here to check if a delta table is V-ORDER optimized.
Duckdb can't read from OneLake abfs either and it doesn't accept onelake_fs object I created above directly. But, we can pass pyarrow dataframe to duckdb thanks to zero-copy integration between the two.
# %pip install duckdb --q #if duckdb is not installed
import sempy.fabric as fabric
import fsspec
import pyarrow.parquet as pq
import duckdb
def get_lakehouse_abfs(lakehouse_name, workspace_name, relative_path=None):
path = mssparkutils.lakehouse.get(name=lakehouse_name, workspaceId = fabric.resolve_workspace_id(workspace_name))['properties']['abfsPath']
if relative_path:
path = path + relative_path
return path
path = get_lakehouse_abfs("product_sales_gold", "Product Sales", relative_path="/Files/covid-19_data.parquet")
#deinf filesystem
filesystem_code = "abfss"
storage_options = {
"account_name": "onelake",
"account_host": "onelake.dfs.fabric.microsoft.com"
}
onelake_fs = fsspec.filesystem(filesystem_code, **storage_options)
#read pyarrow
pq_table = pq.read_table(path, filesystem=onelake_fs)
#zero-copy from pyarrow to duckdb
con = duckdb.connect(database=':memory:')
con.register('covid_data', pq_table)
result = con.execute("SELECT * FROM covid_data LIMIT 5").fetchall()
display(result)
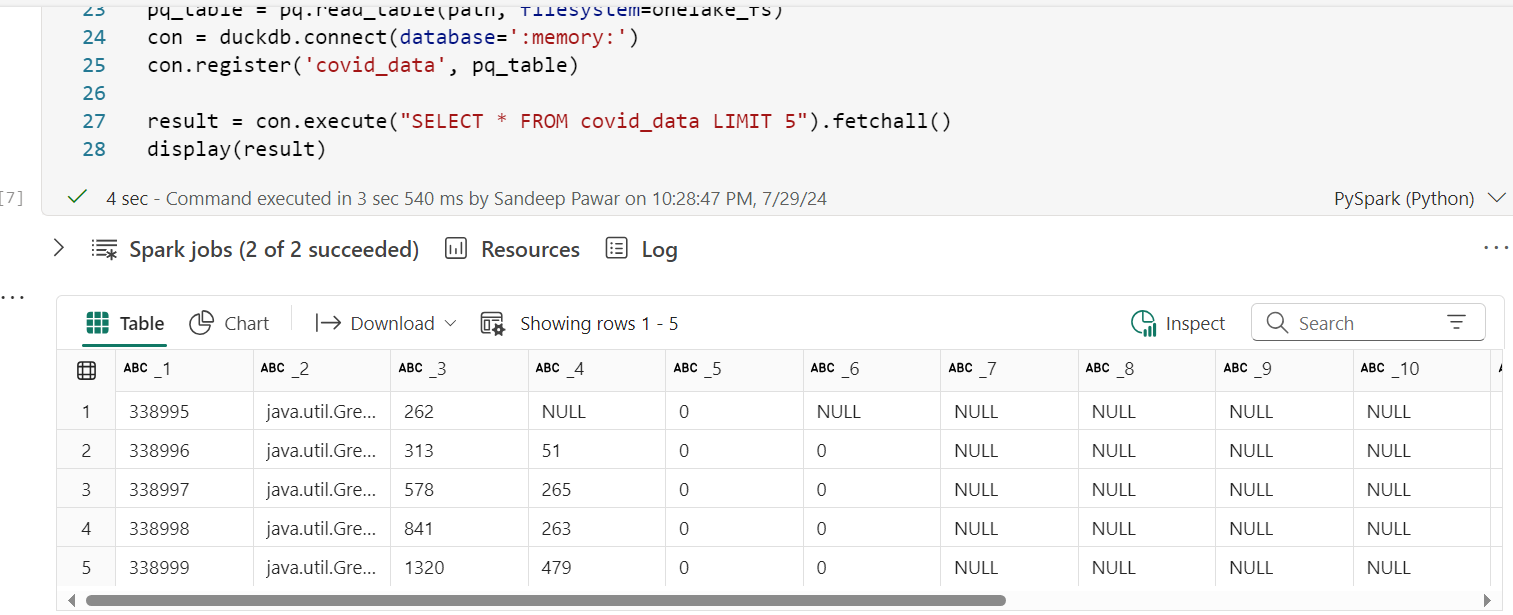
Daft
Daft is an interesting OSS Python project that's gaining a lot of traction. It's a distributed query engine and aims to replace spark. Read how AWS used Ray + Daft to replace spark at exabyte scale. For Azure, you can define the AzureConfig and set use_fabric_endpoint =True as below
AzureConfig(storage_account="onelake",use_fabric_endpoint = True )
But it didn't work for me as it still expects the bearer_token to be passed. But you can create a Daft dataframe from a pyarrow dataframe, so above method works for daft too. Similar to duckdb, Daft has a zero-copy integration with arrow which means, arrow is just used as a medium to point to the data without copying the data twice in memory.
# %pip install getdaft --q
#create a pyrrow table
pq_table = pq.read_table(path, filesystem=onelake_fs)
#create daft df from pyarrow
daft_df = daft.from_arrow(pq_table)
daft_df
This will become super handy when Python notebooks become available in Fabric. Mim talks about duckdb, polars, single node Python notebooks like everyday.
Once you define the local filesystem as above, paths are straight forward: <worskpace_id>/<lakehouse_id>/Files. See below:
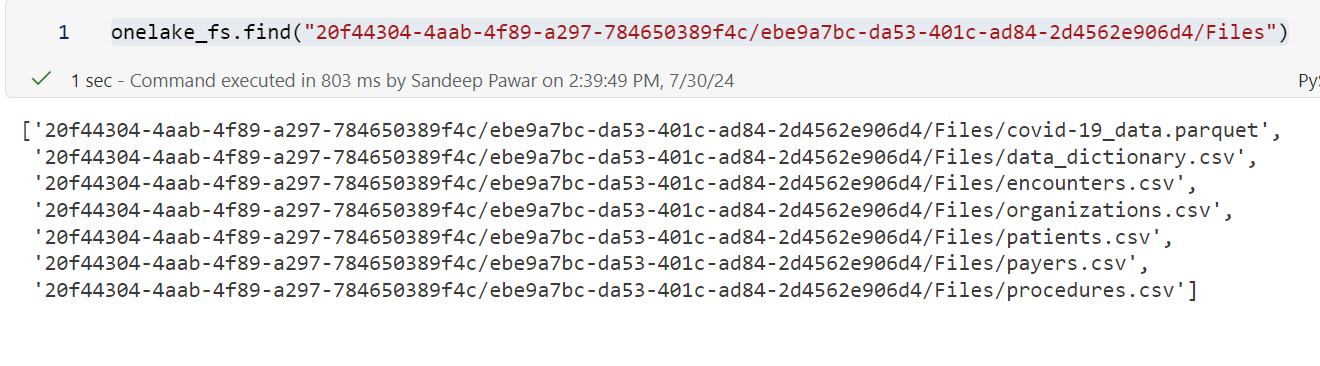
There are other advantages of fsspec but they are not significant in my opinion or not relevant to Fabric. If you are still curious, see the references below.
References
Subscribe to my newsletter
Read articles from Sandeep Pawar directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by

Sandeep Pawar
Sandeep Pawar
Principal Program Manager, Microsoft Fabric CAT helping users and organizations build scalable, insightful, secure solutions. Blogs, opinions are my own and do not represent my employer.