Programming the genome with CRISPR
 freeCodeCamp
freeCodeCamp
By Josh McMenemy
How scientists edit genomes with the help of computers

CRISPR (pronounced “crisper”) is part of a bacterial immune system evolved to ‘remember’ and remove invading viral DNA.
Its name is short for ‘Clustered Regularly Interspaced Short Palindromic Repeats’. But despite its mouthful of an acronym and complex biological origins, its engineering application is straightforward. To get started, there is only one protein you need to understand — Cas9.
Cas9 searches for a specified DNA sequence and cuts it by breaking both strands of the DNA molecule. This protein is useful to researchers because they can ‘program’ it to target any DNA sequence. A sgRNA (‘single guide’ RNA) molecule determines the sequence that Cas9 binds to. RNA is a biological molecule similar to DNA, that can bind to proteins and DNA.
sgRNAs are short sequences with a constant region and variable region. The constant region attaches the sgRNA to the Cas9 protein. The variable region causes Cas9 to bind to the DNA sequence that complements it (see the diagram below).
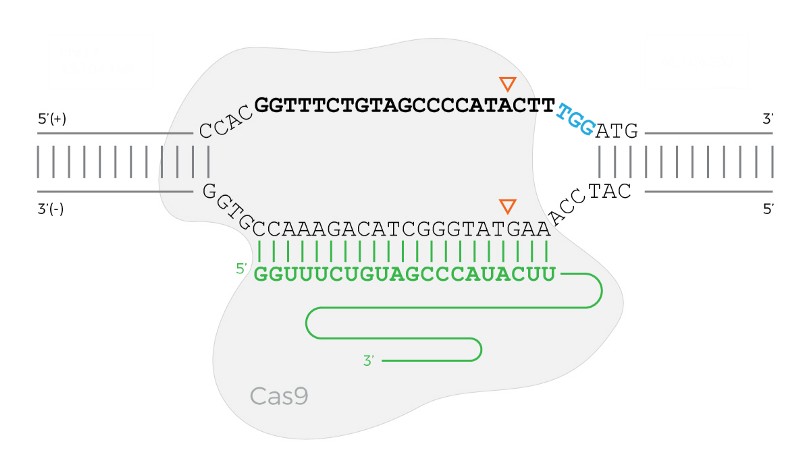 The Cas9 protein bound to the DNA when the PAM sequence is on the forward (top) strand. The bold sequence is the target sequence, the green sequence is the sgRNA, and the three blue characters are the PAM. The triangles show where Cas9 will cut the DNA.
The Cas9 protein bound to the DNA when the PAM sequence is on the forward (top) strand. The bold sequence is the target sequence, the green sequence is the sgRNA, and the three blue characters are the PAM. The triangles show where Cas9 will cut the DNA.
Making sgRNA is cheap and fast. This allows researchers to quickly set up a Cas9 experiment that cuts any DNA sequence. Well, not actually any sequence. There is a small constraint: the target sequence must be flanked by the correct PAM (protospacer adjacent motif) — a short sequence of DNA.
Streptococcus pyogenes is an infectious species of bacteria. In the version of Cas9 it produces, the PAM motif is ‘NGG’, where N is any nucleotide (the ‘letters’ that make up DNA).
Luckily, the motif ‘NGG’ occurs roughly once every 42 basepairs in the human genome. This mean that researchers can find a target site near almost every sequence of interest.
Depending on the experimental set up, these cuts in the DNA can either cause a random change or a precise change to the DNA sequence (more on this later).
Before jumping into writing this program, I recommend studying the Cas9 diagram below.
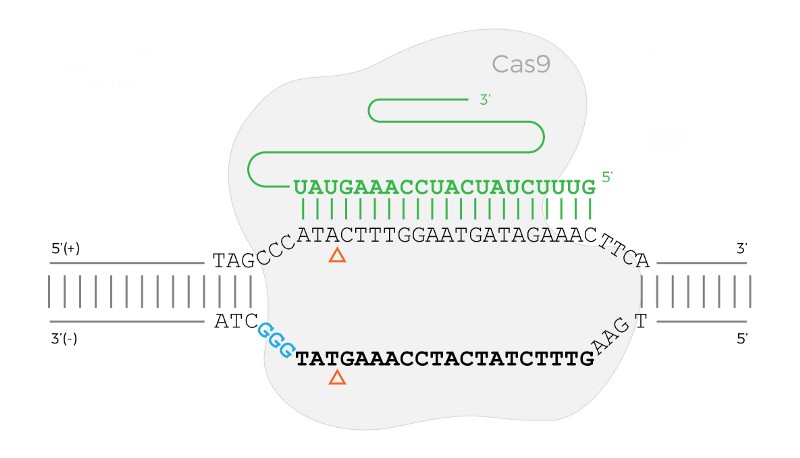 The Cas9 protein bound to a DNA sequence when the PAM sequence is on the reverse (bottom) strand.
The Cas9 protein bound to a DNA sequence when the PAM sequence is on the reverse (bottom) strand.
Note that DNA and RNA have a directionality based on their chemical structure. One end of the molecule is referred to as the 5(‘five-prime’) end, and the other is referred to as the 3 (‘three-prime’) end. This is important, because the sequences 5— AGG — 3 is not the same as 3— AGG — 5.
By convention, DNA and RNA sequences are assumed to be written 5to 3 unless otherwise marked. Sequences read in the 5— 3 direction are called ‘forward’ sequences. Sequences read the other way (3— 5) are called ‘reverse’ sequences. This is an arbitrary convention.
The diagram above shows an example of Cas9 bound when the PAM is on the reverse (bottom) strand.
Your first CRISPR program
The scenario
A scientist has a DNA sequence of interest and wants a list of all CRISPR targets contained in the sequence. Finding every target by hand is tedious and error prone.
The scientist wants a simple program where they can input a DNA sequence and have all possible Cas9 target sites returned. The scientist would also like the cut position and PAM sequence for each target site.
EXAMPLE INPUT (from Figure 1): 'CCACGGTTTCTGTAGCCCCATACTTTGGATG'
EXAMPLE OUTPUT: [{ 'cut_pos': 6, 'pam_seq': 'TGG', 'target_seq': 'GTATGGGGCTACAGAAACCG', 'strand': 'reverse' }, { 'cut_pos': 22, 'pam_seq': 'TGG', 'target_seq': 'GTTTCTGTAGCCCCATACTT', 'strand': 'forward' }]
First, how do we find CRISPR targets in the sequence? Remember that the Cas9 protein can bind anywhere there is a ‘NGG’ motif.
The first step is to loop through the sequence looking for matches. When the program finds a ‘NGG’ match, we want to subtract three positions from the start of the PAM site, since that is where Cas9 cuts the DNA.
Then, we want to record the twenty basepairs before the PAM as the target sequence. Sounds good?
Well, the algorithm described above would actually miss about half of all CRISPR sites — because DNA is double stranded. This means if a ‘CCN’ is the sequence on the forward strand, then ‘NGG’ is the sequence on the reverse strand.
The program must also search for ‘CCN’ using similar logic for the reverse strand.
Example program
Not all CRISPR targets are equal
When CRISPR was first catching on, researchers would often pull up a sequence on their computer and pick targets by hand. Designing the optimal sgRNA has now become much more complex. Below are brief introductions to this complexity.
Off-targets
Researchers soon realized that Cas9 would sometimes bind and cut at loci that did not exactly match the target sequence. These off-target cuts would cause unintended changes in a researcher’s experiment (or potentially a patient’s genome in the case of a therapy!)
To design a good guide, a program must look at the entire genome (which is approximately 3 billion nucleotides for humans) to calculate an off-target score. Researchers have also recently engineered the Cas9 protein to have less off-target activity.
Knockout
When Cas9 binds, it creates a cut by making a double strand break to the DNA molecule. Most of the time, a cell can repair this break through a biochemcial pathway (called non-homologous end joining, or NHEJ).
This pathway is not always perfect, and sometimes when Cas9 cuts, the repair process makes a small insertion or deletion in the DNA sequence. In a protein-coding region of DNA, these small insertions and deletions cause a frameshift mutation — which will often disrupt the protein’s function.
Researchers will often knockout a gene to figure out how a protein affects a specific cell function or phenotype. Creating a knockout edit adds extra constraints to the sgRNA design, because now the guide must land in the coding region of the gene.
Editing
Instead of knocking out a gene, there are many times a scientist wants to make a precision edit. This is especially useful when trying to correct a disease causing a mutation. The best way to do this is still being researched. Most methods involve adding an extra donor piece of DNA.
On-target score
Some sgRNA sequences will cause Cas9 to cut better than others. Researchers have compared cutting efficiency across thousands of Cas9 targets to create predictive models of a sgRNA’s cutting efficiency.
Microsoft even supports an open source repository for ‘Machine Learning-Based Predictive Modeling of CRISPR/Cas9 guide efficiency’.
Other CRISPR-Cas systems
Researchers have discovered CRISPR-Cas systems in other bacteria. These other systems have different PAMs.
Final notes
Hope you learned something new! If you want to learn more about the biology, medical applications, commercial applications, or ethical implications of CRISPR-Cas genome engineering, then I recommend reading A Crack in Creation by Jennifer Doudna and Samuel Sternberg. Jennifer Doudna is one of the original discovers of CRISPR’s underpinnings.
About the Author
I was previously an undergraduate researcher in the Gersbach Lab at Duke University, and I am currently a Software Engineer at a Synthego.
Subscribe to my newsletter
Read articles from freeCodeCamp directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by

freeCodeCamp
freeCodeCamp
Learn to code. Build projects. Earn certifications—All for free.