A CASE STUDY HIGHLIGHTING BugSigDB AND IT’S IMPACT ON THE COMMUNITY
 bolanle wahab
bolanle wahab
About:
BugSigDB.org is an editable Semantic MediaWiki that provides manually curated, expert-reviewed, syntax-standardized reporting of the published literature on microbial signatures. Curators standardize taxa to the NCBI taxonomy and standardize key information about the studies, experiments, and analytical methods that generated these signatures using data entry forms, controlled vocabulary, and ontologies. BugSigDB provides curated published signatures of differentially abundant microbes associated with a wide range of health outcomes, pharmaceutical usage (for example, antibiotics), experiments on animal models, randomized clinical trials, and microbial attributes and is built on the technology of Wikipedia to allow community contributions, revisions and review for quality control.(Geistlinger et al., 2024)
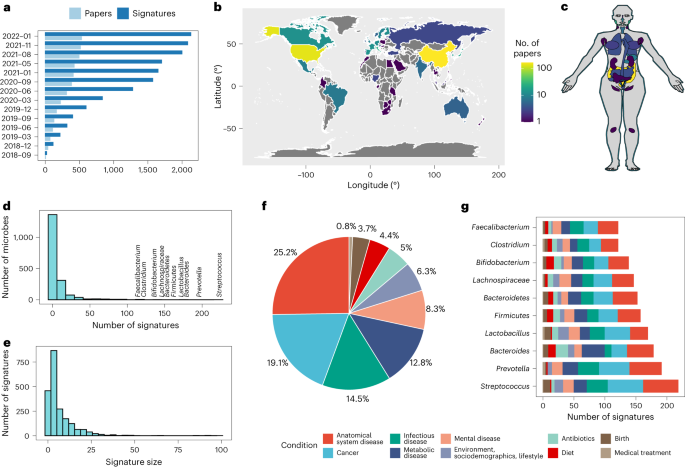
BugSigDB, a curated database of experimentally derived microbial signatures Geistlinger, L., Mirzayi, C., Zohra, F., Azhar, R., Elsafoury, S., Grieve, C., Wokaty, J., David, S., Sengupta, P., Hecht, I., Ravikrishnan, A., Gonçalves, R. S., Franzosa, E., Raman, K., Carey, V., Dowd, J. B., Jones, H. E., Davis, S., Segata, N., . . . Waldron, L. (2024). BugSigDB captures patterns of differential abundance across a broad range of host-associated microbial signatures. Nature Biotechnology, 42(5), 790-802. https://doi.org/10.1038/s41587-023-01872-y
Problem Statement:
The rapid expansion of microbiome research has resulted in vast amounts of data across numerous studies. These issues have limited the potential for large-scale meta-analyses and hindered the discovery of generalizable patterns across different microbiome studies. Researchers face several challenges:
Data Fragmentation: Microbiome data is scattered across many publications, making it difficult to find and compare relevant datasets.
Lack of Standardization: Variations in taxonomy, study design, and reporting methods complicate data aggregation and cross-study comparisons.
Reproducibility Issues: Without standardized data, it is difficult for researchers to reproduce findings, which hinders scientific progress.
Solution:
BugSigDB was developed to address the challenges of fragmented and non-standardized microbiome data by providing a publicly accessible, open-source platform that curates microbiome signatures from published studies. The platform offers the following key features:
Curated Microbiome Signatures: BugSigDB collects and organizes microbiome data from scientific studies, focusing on the relationships between microbial taxa and specific conditions or body sites.
Data Standardization: The platform standardizes metadata, enabling researchers to perform cross-study comparisons and improve reproducibility easily.
Open-Source and Collaborative: As a community-driven project, BugSigDB encourages researchers to contribute new signatures and refine existing data, enhancing collaboration and transparency.
User-Friendly Interface: BugSigDB provides an intuitive interface that allows users to explore signatures based on microbial taxa, body sites, study designs, and conditions, making it an indispensable tool for comparative analysis in microbiome research.
Success Factors:
Open Source and Community-Driven: BugSigDB’s open-source nature allows for contributions from the global research community. The platform's collaborative environment ensures that it remains up-to-date and relevant.
User-Friendly Interface: BugSigDB’s design prioritizes ease of use to allow researchers, regardless of technical expertise, to access and analyze microbiome data.
Rigorous Curation Process: Each signature added to BugSigDB undergoes a stringent curation process to ensure the accuracy and reliability of the data.
Support for Integration with Other Tools: BugSigDB provides resources for researchers to integrate its data with other bioinformatics tools and databases to improve its utility in large-scale analysis and research projects.
Conclusion:
BugSigDB has emerged as a vital resource in the field of microbiome research. Its ability to aggregate, standardize, and disseminate microbiome signatures has significantly impacted how researchers approach the study of microbial communities. The process of reducing data fragmentation, standardizing metadata, and improving accessibility has made it a valuable tool for advancing microbiome research, setting a benchmark for future open-source scientific databases.
Subscribe to my newsletter
Read articles from bolanle wahab directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
