Hierarchical Navigable Small World (HNSW)
 Sai Prasanna Maharana
Sai Prasanna MaharanaCertainly!
Hierarchical Navigable Small World (HNSW) is a popular algorithm used in vector databases for efficient approximate nearest neighbor (ANN) search in high-dimensional spaces. It is particularly effective for large-scale datasets due to its scalability and performance. When combined with cosine similarity as the distance metric, HNSW enables fast and accurate retrieval of vectors based on their angular similarity.
Table of Contents
Introduction to HNSW
Understanding Cosine Similarity
How HNSW Works with Cosine Similarity
Building the HNSW Graph
Navigating the Graph for Search
Implementation Steps
Index Construction
Query Processing
Code Example Using FAISS
Advantages of HNSW with Cosine Similarity
Considerations and Best Practices
Conclusion
References
1. Introduction to HNSW
HNSW (Hierarchical Navigable Small World) is an algorithm for approximate nearest neighbor search based on the concept of navigable small-world graphs. It constructs a multi-layer graph where each layer represents a network of connections between data points (vectors). The algorithm efficiently navigates through these layers to find the nearest neighbors of a query vector.
Key Features:
Hierarchical Structure: Multiple layers with decreasing levels of abstraction.
Navigability: Efficient traversal using greedy search strategies.
Scalability: Handles millions of vectors with high performance.
2. Understanding Cosine Similarity
Cosine similarity measures the cosine of the angle between two vectors in a multi-dimensional space. It is defined as:
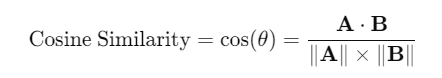
Range: The value ranges from -1 to 1.
Interpretation:
1: Vectors are identical in direction.
0: Vectors are orthogonal (no similarity).
-1: Vectors are diametrically opposed.
Why Use Cosine Similarity?
Scale Invariance: Focuses on the orientation rather than magnitude.
Common in NLP: Frequently used in text embeddings where direction matters more than magnitude.
3. How HNSW Works with Cosine Similarity
a. Building the HNSW Graph
Initialization:
Start with an empty graph.
Determine the maximum layer
Lmaxbased on the logarithm of the number of elements.
Insertion of Nodes:
Random Level Assignment: Each new vector is assigned a random maximum layer
L(up toLmax) following an exponential distribution.Connecting Nodes:
For each layer from
Lmaxdown to level 0:Greedy Search: Find the closest nodes in the current layer to the new vector using cosine similarity.
Establish Connections: Connect the new vector to these nodes, forming edges in the graph.
Neighbor Selection: Limit the number of connections (controlled by parameter
M) to balance graph density and search speed.
b. Navigating the Graph for Search
Starting Point:
- Begin the search from an entry point at the top layer (
Lmax).
- Begin the search from an entry point at the top layer (
Greedy Search:
At each layer:
Current Node: Start from the current node.
Neighbors Exploration: Examine the neighbors of the current node.
Distance Calculation: Compute cosine similarity between the query vector and neighboring nodes.
Move Decision: Move to the neighbor with the highest cosine similarity (closest in angular distance).
Layer Transition:
If no closer neighbor is found at the current layer, descend to the next lower layer.
Repeat the greedy search at each layer until reaching layer 0.
Final Search at Layer 0:
- Perform a more thorough search (e.g., beam search) to refine the nearest neighbors.
4. Implementation Steps
a. Index Construction
Data Preparation: Normalize all vectors to unit length to use cosine similarity effectively.
import numpy as np def normalize_vectors(vectors): norms = np.linalg.norm(vectors, axis=1, keepdims=True) return vectors / normsGraph Building: Use an HNSW library (e.g., FAISS, Annoy, hnswlib) to build the index.
b. Query Processing
Query Normalization: Normalize the query vector to unit length.
Similarity Search: Use the HNSW index to find the nearest neighbors based on cosine similarity.
5. Code Example Using FAISS
Let's demonstrate how HNSW works with cosine similarity using the FAISS library.
a. Install FAISS
pip install faiss-cpu # For CPU
# or
pip install faiss-gpu # For GPU
b. Sample Dataset
import numpy as np
# Generate a sample dataset of 10,000 128-dimensional vectors
np.random.seed(42)
data = np.random.random((10000, 128)).astype('float32')
c. Normalize Vectors
def normalize_vectors(vectors):
norms = np.linalg.norm(vectors, axis=1, keepdims=True)
return vectors / norms
data_normalized = normalize_vectors(data)
d. Build the HNSW Index
import faiss
d = data.shape[1] # Dimension of vectors
# Use Inner Product (Dot Product) for Cosine Similarity
index = faiss.index_factory(d, "HNSW32", faiss.METRIC_INNER_PRODUCT)
# Set HNSW parameters (optional)
hnsw_param = index.hnsw
hnsw_param.nb_neighbors = 32 # M parameter
hnsw_param.efConstruction = 200 # Controls index time and accuracy
# Train the index if necessary (not required for HNSW with flat vectors)
# Add vectors to the index
index.add(data_normalized)
e. Perform a Search
# Normalize the query vector
query_vector = np.random.random((1, 128)).astype('float32')
query_vector_normalized = normalize_vectors(query_vector)
# Set search parameters
index.hnsw.efSearch = 50 # Controls search speed vs. accuracy
# Perform the search
k = 5 # Number of nearest neighbors
distances, indices = index.search(query_vector_normalized, k)
# Display results
print("Nearest neighbors (indices):", indices)
print("Distances (cosine similarity):", distances)
Note: In FAISS, when using METRIC_INNER_PRODUCT, the distances returned are the inner product values. Since the vectors are normalized, the inner product is equivalent to cosine similarity.
f. Explanation
Normalization: By normalizing vectors to unit length, the inner product becomes cosine similarity.
Index Creation: The
HNSW32specifies that each node in the graph will have up to 32 connections.Search Parameters:
efSearchcontrols the trade-off between search speed and recall.
6. Advantages of HNSW with Cosine Similarity
Efficient Retrieval: HNSW provides sub-linear query time complexity, making it suitable for large datasets.
High Recall: Capable of achieving high recall rates with appropriate tuning.
Scalability: Can handle millions of vectors with reasonable memory consumption.
Flexibility: Supports various distance metrics, including cosine similarity.
7. Considerations and Best Practices
Vector Normalization:
Essential when using cosine similarity to ensure accurate distance computations.
Normalization should be applied consistently to both index vectors and query vectors.
Parameter Tuning:
M (nb_neighbors): Controls the graph's connectivity. Higher values improve accuracy but increase memory usage.
efConstruction: Affects the quality of the index during construction. Higher values yield better recall.
efSearch: Adjusts the search depth. Increasing
efSearchimproves recall at the expense of query time.
Memory Consumption:
HNSW indexes can consume significant memory, especially with large
Mvalues.Monitor memory usage and adjust parameters accordingly.
Distance Metric Consistency:
Ensure that the distance metric used during index construction matches the metric used during search.
In FAISS, use
METRIC_INNER_PRODUCTwith normalized vectors for cosine similarity.
Library Selection:
- Choose a library that supports HNSW and cosine similarity, such as FAISS or hnswlib.
8. Conclusion
HNSW is a powerful algorithm for approximate nearest neighbor search in vector databases. When combined with cosine similarity, it enables efficient and accurate retrieval of vectors based on their angular relationships. By constructing a hierarchical graph and navigating it using greedy search strategies, HNSW provides a scalable solution suitable for production environments handling large-scale, high-dimensional data.
9. References
HNSW Paper: Efficient and robust approximate nearest neighbor search using Hierarchical Navigable Small World graphs
FAISS Library: FAISS - A library for efficient similarity search
Cosine Similarity: Understanding Cosine Similarity
Subscribe to my newsletter
Read articles from Sai Prasanna Maharana directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
