Applying OASIS-3 Data to TFF
 KiwiChip
KiwiChiphttps://arxiv.org/abs/2112.05761
Pre-processing codes
preprocessing.py
import os
import numpy as np
import nibabel as nib
import torch
from multiprocessing import Process, Queue
def read_hcp(file_path,global_norm_path,per_voxel_norm_path,hand,count,queue=None):
img_orig = torch.from_numpy(np.asanyarray(nib.load(file_path).dataobj)[8:-8, 8:-8, :-10, 10:]).to(dtype=torch.float32)
background = img_orig == 0
img_temp = (img_orig - img_orig[~background].mean()) / (img_orig[~background].std())
img = torch.empty(img_orig.shape)
img[background] = img_temp.min()
img[~background] = img_temp[~background]
img = torch.split(img, 1, 3)
for i, TR in enumerate(img):
torch.save(TR.clone(),
os.path.join(global_norm_path, 'rfMRI_' + hand + '_TR_' + str(i) + '.pt'))
# repeat for per voxel normalization
img_temp = (img_orig - img_orig.mean(dim=3, keepdims=True)) / (img_orig.std(dim=3, keepdims=True))
img = torch.empty(img_orig.shape)
img[background] = img_temp.min()
img[~background] = img_temp[~background]
img = torch.split(img, 1, 3)
for i, TR in enumerate(img):
torch.save(TR.clone(),
os.path.join(per_voxel_norm_path, 'rfMRI_' + hand + '_TR_' + str(i) + '.pt'))
print('finished another subject. count is now {}'.format(count))
def main():
hcp_path = r'D:\users\Gony\HCP-1200'
all_files_path = os.path.join(hcp_path,'extract_S1200_data')
queue = Queue()
count = 0
for subj in os.listdir(all_files_path):
subj_path = os.path.join(all_files_path,subj)
try:
file_path = os.path.join(subj_path,os.listdir(subj_path)[0])
hand = file_path[file_path.find('REST1_')+6:file_path.find('.nii')]
global_norm_path = os.path.join(hcp_path,'MNI_to_TRs',subj,'global_normalize')
per_vox_norm_path = os.path.join(hcp_path, 'MNI_to_TRs', subj, 'per_voxel_normalize')
os.makedirs(global_norm_path, exist_ok=True)
os.makedirs(per_vox_norm_path, exist_ok=True)
count += 1
print('start working on subject '+ subj)
p = Process(target=read_hcp, args=(file_path,global_norm_path,per_vox_norm_path,hand,count, queue))
p.start()
if count % 20 == 0:
p.join() # this blocks until the process terminates
except Exception:
print('encountered problem with '+subj)
print(Exception)
if __name__ == '__main__':
main()
The read_hcp function is central to the process, reading fMRI images from HCP data, normalizing them in two ways, and saving them as tensor files for each time resolution (TR).
1. Data Loading and Slicing
Load fMRI data in
.niiformat using thenibabellibrary.Trim unnecessary parts of the data (
img_orig = torch.from_numpy(np.asanyarray(nib.load(file_path).dataobj)[8:-8, 8:-8, :-10, 10:])) to focus on the region of interest.
2. Background Masking
- Create a mask using
background = img_orig == 0to distinguish between the background and actual data areas.
3. Normalization
Global Normalization: Calculate the mean and standard deviation of the data excluding the background, and normalize based on these values.
img_temp = (img_orig - img_orig[~background].mean()) / (img_orig[~background].std())Assign the minimum value to the background area to differentiate it from the data in the normalized image (
img).Voxel-wise Normalization: Normalize each voxel by calculating the mean and standard deviation along the time axis (
dim=3).img_temp = (img_orig - img_orig.mean(dim=3, keepdims=True)) / (img_orig.std(dim=3, keepdims=True))
4. Splitting and Saving by Time Resolution (TR)
Split the normalized image by time resolution using
torch.split(img, 1, 3)and save each as a.ptfile.Save the results of global and voxel-wise normalization in separate files, creating
.ptfiles for each TR to be loaded individually during model training.
5. Multiprocessing
- Use
Processfor parallel processing when handling multiple subjects, and manage memory by cleaning up processes withp.join()every 20 datasets.
In summary, this code performs preprocessing in the order of data slicing → background masking → two types of normalization (global and voxel-wise) → splitting by TR and saving as .pt files.
→ Since OASIS-3 data is already preprocessed, adjustments will be made to check data format and dimensions, and modify the data loader section.
dataloader.py
Responsible for loading datasets and creating training, validation, and test sets, returning them as data loaders.
import numpy as np
from torch.utils.data import DataLoader,Subset
from data_preprocess_and_load.datasets import *
from utils import reproducibility
class DataHandler():
def __init__(self,test=False,**kwargs):
self.test = test
self.kwargs = kwargs
self.dataset_name = kwargs.get('dataset_name')
self.splits_folder = Path(kwargs.get('base_path')).joinpath('splits',self.dataset_name)
self.splits_folder.mkdir(exist_ok=True)
self.seed = kwargs.get('seed')
self.current_split = self.splits_folder.joinpath('seed_{}.txt'.format(self.seed))
def get_dataset(self):
if self.dataset_name == 'S1200':
return rest_1200_3D
elif self.dataset_name == 'ucla':
return ucla
else:
raise NotImplementedError
def current_split_exists(self):
return self.current_split.exists()
def create_dataloaders(self):
reproducibility(**self.kwargs)
dataset = self.get_dataset()
train_loader = dataset(**self.kwargs)
eval_loader = dataset(**self.kwargs)
eval_loader.augment = None
self.subject_list = train_loader.index_l
if self.current_split_exists():
train_names, val_names, test_names = self.load_split()
train_idx, val_idx, test_idx = self.convert_subject_list_to_idx_list(train_names,val_names,test_names,self.subject_list)
else:
train_idx,val_idx,test_idx = self.determine_split_randomly(self.subject_list,**self.kwargs)
# train_idx = [train_idx[x] for x in torch.randperm(len(train_idx))[:10]]
# val_idx = [val_idx[x] for x in torch.randperm(len(val_idx))[:10]]
train_loader = Subset(train_loader, train_idx)
val_loader = Subset(eval_loader, val_idx)
test_loader = Subset(eval_loader, test_idx)
training_generator = DataLoader(train_loader, **self.get_params(**self.kwargs))
val_generator = DataLoader(val_loader, **self.get_params(eval=True,**self.kwargs))
test_generator = DataLoader(test_loader, **self.get_params(eval=True,**self.kwargs)) if self.test else None
return training_generator, val_generator, test_generator
def get_params(self,eval=False,**kwargs):
batch_size = kwargs.get('batch_size')
workers = kwargs.get('workers')
cuda = kwargs.get('cuda')
if eval:
workers = 0
params = {'batch_size': batch_size,
'shuffle': True,
'num_workers': workers,
'drop_last': True,
'pin_memory': False, # True if cuda else False,
'persistent_workers': True if workers > 0 and cuda else False}
return params
def save_split(self,sets_dict):
with open(self.current_split,'w+') as f:
for name,subj_list in sets_dict.items():
f.write(name + '\n')
for subj_name in subj_list:
f.write(str(subj_name) + '\n')
def convert_subject_list_to_idx_list(self,train_names,val_names,test_names,subj_list):
subj_idx = np.array([str(x[0]) for x in subj_list])
train_idx = np.where(np.in1d(subj_idx, train_names))[0].tolist()
val_idx = np.where(np.in1d(subj_idx, val_names))[0].tolist()
test_idx = np.where(np.in1d(subj_idx, test_names))[0].tolist()
return train_idx,val_idx,test_idx
def determine_split_randomly(self,index_l,**kwargs):
train_percent = kwargs.get('train_split')
val_percent = kwargs.get('val_split')
S = len(np.unique([x[0] for x in index_l]))
S_train = int(S * train_percent)
S_val = int(S * val_percent)
S_train = np.random.choice(S, S_train, replace=False)
remaining = np.setdiff1d(np.arange(S), S_train)
S_val = np.random.choice(remaining,S_val, replace=False)
S_test = np.setdiff1d(np.arange(S), np.concatenate([S_train,S_val]))
train_idx,val_idx,test_idx = self.convert_subject_list_to_idx_list(S_train,S_val,S_test,self.subject_list)
self.save_split({'train_subjects':S_train,'val_subjects':S_val,'test_subjects':S_test})
return train_idx,val_idx,test_idx
def load_split(self):
subject_order = open(self.current_split, 'r').readlines()
subject_order = [x[:-1] for x in subject_order]
train_index = np.argmax(['train' in line for line in subject_order])
val_index = np.argmax(['val' in line for line in subject_order])
test_index = np.argmax(['test' in line for line in subject_order])
train_names = subject_order[train_index + 1:val_index]
val_names = subject_order[val_index+1:test_index]
test_names = subject_order[test_index + 1:]
return train_names,val_names,test_names
Key Code Analysis
Dataset Selection (
get_datasetmethod):Selects the dataset class based on
self.dataset_name. Returnsrest_1200_3Dfor'S1200'anduclafor'ucla'.To adapt this code for the OASIS-3 dataset, modify the
get_dataset()method to return a class or function that handles OASIS-3 data.
Dataset Splitting (
create_dataloadersmethod):Checks for existing dataset split files with
self.current_split_exists(), and loads them withload_split()if they exist.If not, uses
determine_split_randomly()to randomly split into training, validation, and test sets, saving the split information withsave_split().
Using the
SubsetClass:- Wraps the split indices in
Subsetto createtrain_loader,val_loader, andtest_loader, which deliver data in batches to the model.
- Wraps the split indices in
Data Loader Configuration (
get_paramsmethod):- Configures data loaders based on batch size, number of workers, and CUDA usage. Optimizes data loading speed with
pin_memoryandpersistent_workersoptions if using CUDA.
- Configures data loaders based on batch size, number of workers, and CUDA usage. Optimizes data loading speed with
Data Loader Return:
- Returns
training_generator,val_generator, andtest_generator, which deliver training, validation, and test data to the model in theTrainerclass.
- Returns
Applying OASIS-3 Data
Add Dataset Class:
- Define a class for handling OASIS-3 data in the
data_preprocess_and_load/datasets.pyfile, and modify theget_dataset()method to return this class.
- Define a class for handling OASIS-3 data in the
Modify Split Criteria:
- Adjust
determine_split_randomly()andconvert_subject_list_to_idx_list()methods to generate indices based on OASIS-3's unique identifiers (e.g., sub-XXXX).
- Adjust
Ensure Data Structure for
TrainerClass:The
forward_pass()method in theTrainerclass passesinput_dict['fmri_sequence']to the model, so OASIS-3 data must be structured similarly.Ensure
input_dictcontains a key like'fmri_sequence'and restructure data to match the expected dimensions (e.g.,[batch_size, channels, depth, height, width]).batch_size: Number of samples in a mini-batchchannels: Number of channels per scandepth: Depth of brain images along the z-axis (e.g., number of slices)height,width: Height and width of each slice
1. File Loading and Dimension Checking
- Load
.niifiles using thenibabellibrary and check data dimensions.
- Load
import nibabel as nib
# .nii 파일 로드
file_path = 'path/to/your/data.nii'
img = nib.load(file_path)
data = img.get_fdata()
print("Data shape:", data.shape)
Interpreting Dimension Check Results:
fMRI data is typically in
[depth, height, width, time]format, wheredepth,height, andwidthare spatial dimensions, andtimeis the TR (time resolution) axis.If the result is
(64, 64, 36, 120), it means the fMRI data consists ofdepth=64,height=64,width=36, andtime=120.
2. Convert to Model Input Format
- Restructure data to match the model's input format,
[batch_size, channels, depth, height, width]or[batch_size, time, depth, height, width].
import torch
# 데이터 차원 변환 (예: [time, depth, height, width] → [time, 1, depth, height, width])
data = torch.from_numpy(data).permute(3, 0, 1, 2).unsqueeze(1) # [time, 1, depth, height, width]
print("Transformed shape:", data.shape)
permute(3, 0, 1, 2)converts[depth, height, width, time]to[time, depth, height, width], andunsqueeze(1)adds a channel dimension to create[time, 1, depth, height, width].
3. Prepare Data for Storage and Use
- Prepare the transformed data to be passed to the model as
fmri_sequence. If you want to save it in.ptformat for later use, you can usetorch.save().
# 변환된 데이터 저장
torch.save(data, 'path/to/transformed_data.pt')
4. Loading Data in the Trainer Class
- If saved as
.ptfiles, modify the data loading section in theTrainerclass to load.ptfiles withtorch.load()and pass them asfmri_sequence.
Model Architecture
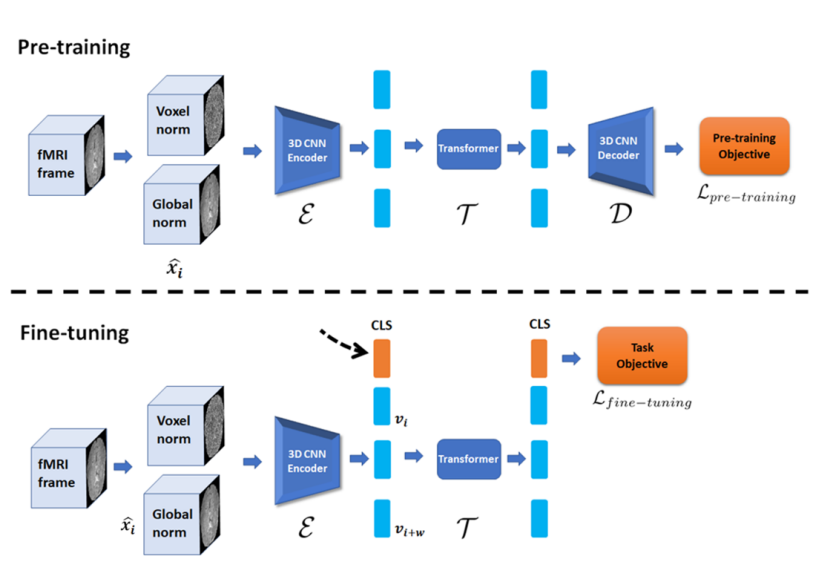
1. Pre-training Phase
- In the pre-training phase, fMRI data is normalized using Voxel norm and Global norm, then reconstructed through a 3D CNN encoder and transformer.
Related Code:
AutoEncoderClassclass AutoEncoder(BaseModel): def __init__(self, dim, **kwargs): super(AutoEncoder, self).__init__() self.task = 'autoencoder_reconstruction' # 3D CNN 인코더 self.encoder = Encoder(**kwargs) self.determine_shapes(self.encoder, dim) # 인코더 차원 결정 kwargs['shapes'] = self.shapes # Bottleneck을 통해 Transformer에 입력 self.into_bert = BottleNeck_in(**kwargs) # Transformer (BERT 모델 사용) self.transformer = Transformer_Block(self.BertConfig, **kwargs) # Bottleneck에서 다시 3D CNN 디코더 입력으로 변환 self.from_bert = BottleNeck_out(**kwargs) # 3D CNN 디코더 self.decoder = Decoder(**kwargs) def forward(self, x): batch_size, Channels_in, W, H, D, T = x.shape x = x.permute(0, 5, 1, 2, 3, 4).reshape(batch_size * T, Channels_in, W, H, D) # 3D CNN 인코더 encoded = self.encoder(x) # Bottleneck (인코더 → 트랜스포머 입력 차원) encoded = self.into_bert(encoded) # Transformer (시간적 특징 학습) encoded = self.transformer(encoded)['sequence'] # Bottleneck (트랜스포머 → 디코더 입력 차원) encoded = self.from_bert(encoded) # 3D CNN 디코더 reconstructed_image = self.decoder(encoded) reconstructed_image = reconstructed_image.reshape(batch_size, T, self.outChannels, W, H, D).permute(0, 2, 3, 4, 5, 1) return {'reconstructed_fmri_sequence': reconstructed_image}
3D CNN Encoder (
EncoderClass):class Encoder(BaseModel): def __init__(self, **kwargs): super(Encoder, self).__init__() self.register_vars(**kwargs) # 첫 번째 다운샘플링 블록 self.down_block1 = nn.Sequential(OrderedDict([ ('conv0', nn.Conv3d(self.inChannels, self.model_depth, kernel_size=3, stride=1, padding=1)), ('sp_drop0', nn.Dropout3d(self.dropout_rates['input'])), ('green0', GreenBlock(self.model_depth, self.model_depth, self.dropout_rates['green'])), ('downsize_0', nn.Conv3d(self.model_depth, self.model_depth * 2, kernel_size=3, stride=2, padding=1))])) # 두 번째 다운샘플링 블록 self.down_block2 = nn.Sequential(OrderedDict([ ('green10', GreenBlock(self.model_depth * 2, self.model_depth * 2, self.dropout_rates['green'])), ('downsize_1', nn.Conv3d(self.model_depth * 2, self.model_depth * 4, kernel_size=3, stride=2, padding=1))])) # 세 번째 다운샘플링 블록 self.down_block3 = nn.Sequential(OrderedDict([ ('green20', GreenBlock(self.model_depth * 4, self.model_depth * 4, self.dropout_rates['green'])), ('downsize_2', nn.Conv3d(self.model_depth * 4, self.model_depth * 8, kernel_size=3, stride=2, padding=1))])) # 최종 특징 추출 블록 self.final_block = nn.Sequential(OrderedDict([ ('green30', GreenBlock(self.model_depth * 8, self.model_depth * 8, self.dropout_rates['green']))])) def forward(self, x): x = self.down_block1(x) x = self.down_block2(x) x = self.down_block3(x) x = self.final_block(x) return xThis
Encoderclass represents the part labeled as3D CNN Encoderin the diagram, and it spatially reduces fMRI data to extract high-dimensional features.Transformer Block (
Transformer_Blockclass):class Transformer_Block(BertPreTrainedModel, BaseModel): def __init__(self, config, **kwargs): super(Transformer_Block, self).__init__(config) self.register_vars(**kwargs) self.cls_pooling = True self.bert = BertModel(self.BertConfig, add_pooling_layer=self.cls_pooling) self.cls_embedding = nn.Sequential(nn.Linear(self.BertConfig.hidden_size, self.BertConfig.hidden_size), nn.LeakyReLU()) def forward(self, x): # Transformer 입력 inputs_embeds = self.cls_embedding(x) outputs = self.bert(inputs_embeds=inputs_embeds) sequence_output = outputs[0][:, 1:, :] pooled_cls = outputs[1] return {'sequence': sequence_output, 'cls': pooled_cls}The part corresponding to the Transformer in the diagram learns temporal patterns.
3D CNN Decoder (
Decoderclass):class Decoder(BaseModel): def __init__(self, **kwargs): super(Decoder, self).__init__() self.register_vars(**kwargs) self.decode_block = nn.Sequential(OrderedDict([ ('upgreen0', UpGreenBlock(self.model_depth * 8, self.model_depth * 4, self.shapes['dim_2'], self.dropout_rates['Up_green'])), ('upgreen1', UpGreenBlock(self.model_depth * 4, self.model_depth * 2, self.shapes['dim_1'], self.dropout_rates['Up_green'])), ('blue_block', nn.Conv3d(self.model_depth, self.model_depth, kernel_size=3, stride=1, padding=1)), ('output_block', nn.Conv3d(in_channels=self.model_depth, out_channels=self.outChannels, kernel_size=1, stride=1)) ])) def forward(self, x): x = self.decode_block(x) return xIn the diagram, the 3D CNN Decoder section corresponds to the
Decoderclass, which reconstructs the original fMRI data form based on features extracted by theEncoder.
2. Fine-tuning Phase
- In the fine-tuning phase, the pre-trained encoder and transformer are used for new prediction tasks (e.g., gender classification).
- A new
CLStoken is added to summarize the features of the entire sequence, which is then passed to the prediction head for making predictions.
Related code:
Encoder_Transformer_finetuneClassclass Encoder_Transformer_finetune(BaseModel): def __init__(self, dim, **kwargs): super(Encoder_Transformer_finetune, self).__init__() self.task = kwargs.get('fine_tune_task') # Pre-trained 3D CNN 인코더 사용 self.encoder = Encoder(**kwargs) self.determine_shapes(self.encoder, dim) kwargs['shapes'] = self.shapes # Transformer 입력을 위한 Bottleneck self.into_bert = BottleNeck_in(**kwargs) # Pre-trained Transformer 사용 self.transformer = Transformer_Block(self.BertConfig, **kwargs) # Fine-tuning을 위한 분류 또는 회귀 헤드 if kwargs.get('fine_tune_task') == 'regression': self.final_activation_func = nn.LeakyReLU() elif kwargs.get('fine_tune_task') == 'binary_classification': self.final_activation_func = nn.Sigmoid() self.label_num = 1 self.regression_head = nn.Sequential(nn.Linear(self.BertConfig.hidden_size, self.label_num), self.final_activation_func) def forward(self, x): batch_size, inChannels, W, H, D, T = x.shape x = x.permute(0, 5, 1, 2, 3, 4).reshape(batch_size * T, inChannels, W, H, D) # 3D CNN 인코더를 통해 특징 추출 encoded = self.encoder(x) # Transformer 입력을 위한 Bottleneck 적용 encoded = self.into_bert(encoded) encoded = encoded.reshape(batch_size, T, -1) # Transformer 통과 (CLS 토큰 포함) transformer_dict = self.transformer(encoded) CLS = transformer_dict['cls'] # Fine-tuning을 위한 분류 또는 회귀 예측 prediction = self.regression_head(CLS) return {self.task: prediction}Fine-tuning Task Head (
regression_head):The
regression_headis the final prediction layer for new tasks during the fine-tuning phase.If
fine_tune_taskis binary classification, Sigmoid is used for binary classification, and if it is regression, LeakyReLU is used.
Architecture Summary
Pre-training Phase: The
AutoEncoderclass, consisting ofEncoder,Transformer_Block, andDecoder, is used to learn the reconstruction of fMRI data with a 3D CNN encoder, transformer, and 3D CNN decoder.Fine-tuning Phase: The
Encoder_Transformer_finetuneclass utilizes the pre-trained encoder and transformer to perform final prediction tasks, summarizing sequence information with a newCLStoken to make the final prediction.
Subscribe to my newsletter
Read articles from KiwiChip directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by

KiwiChip
KiwiChip
I'm currently learning Python and studying RAG (Retrieval-Augmented Generation).