Predicting Leukemia Types Using Gene Expression Data and Logistic Regression
 Gaurav Bhatnagar
Gaurav BhatnagarIntroduction
Leukemia, a cancer affecting blood and bone marrow, can manifest in different forms. Two of the most prevalent types are Acute Lymphoblastic Leukemia (ALL) and Acute Myeloid Leukemia (AML). Accurately classifying these two types is crucial for treatment and prognosis. Traditionally, this process is complex, requiring expert interpretation. But can we use machine learning to classify leukemia types based on gene expression data?
In this blog, I'll walk you through how I developed a Logistic Regression model to classify ALL and AML using gene expression profiles. We’ll cover the steps from data loading and preprocessing, to model evaluation, and the results we achieved.
Problem Statement
The goal of this project is to develop a machine learning model capable of classifying leukemia types using gene expression data. We’ll focus on Logistic Regression due to its simplicity and effectiveness with binary classification problems like ALL vs. AML. This project could have real-world implications for speeding up leukemia diagnosis and improving clinical outcomes.
Methodology
Data Loading and Preprocessing
Filtering: We remove columns containing irrelevant information such as "call" columns.
Cleaning: Handle any missing data and ensure the dataset is consistent.
Scaling: Apply StandardScaler to normalize the gene expression data so all features are on the same scale, which is critical for ensuring the model performs optimally.
Dimensionality Reduction
With over 7,000 genes in the dataset, training a model on such a high-dimensional dataset could lead to overfitting. To address this, we use Principal Component Analysis (PCA) to reduce the dataset down to the most essential components.
PCA helps us condense the data while retaining 70% of the variance. This step reduced the feature set to just 15 components, making the model training process more efficient.
Model Training and Hyperparameter Tuning
Now comes the exciting part: training the model. Logistic Regression was chosen for its ability to handle binary classification tasks like predicting whether a patient has ALL or AML. Additionally, Logistic Regression models are interpretable, making it easier to understand the importance of different features in the decision-making process.
But how do we ensure the model is well-tuned? Enter GridSearchCV, a tool for hyperparameter tuning. This process involved trying out different combinations of parameters (like the regularization strength
Cand penalty type) to identify the optimal settings.After tuning, the best parameters for the Logistic Regression model were:
C: 0.01penalty: L2 (Ridge regularization)
Model Evaluation
To assess how well the model performs, we used multiple evaluation metrics on an independent test dataset of 34 patients. These metrics include:
Accuracy: How often the model predicts the correct class.
Precision: The proportion of true positive predictions out of all positive predictions.
F1 Score: A balance between precision and recall, ideal for imbalanced datasets.
Confusion Matrix: A breakdown of true positives, true negatives, false positives, and false negatives, providing deeper insights into the model's performance.
Results
Accuracy: 94.1%
Precision: 92.9%
F1 Score: 92.9%
Confusion Matrix:
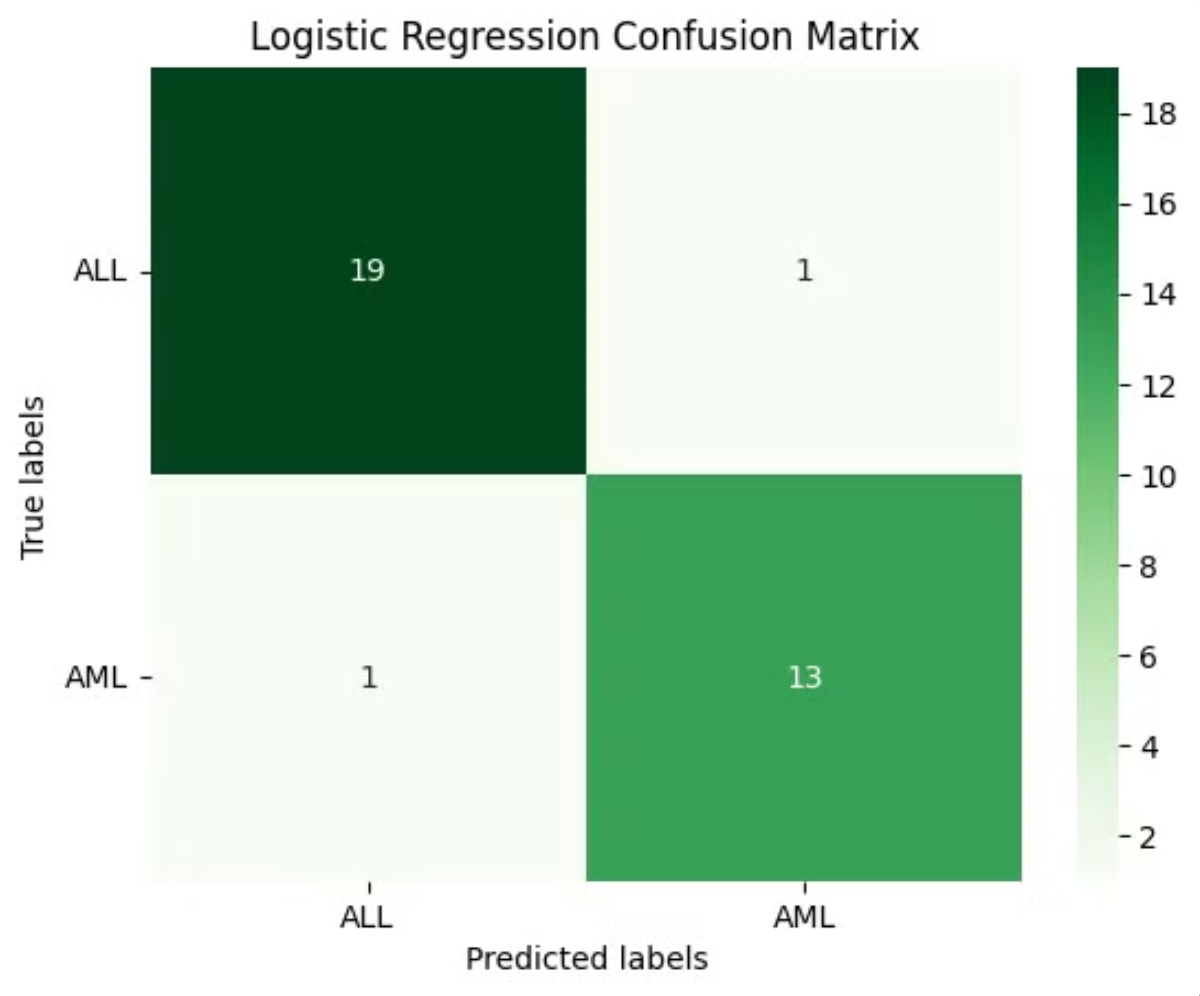
Key Insights and Takeaways
This project demonstrates the potential of machine learning in healthcare, specifically in leukemia classification. here are some key insights:
Data Preprocessing: Ensuring that the data is properly cleaned, scaled, and transformed is critical to the success of the model.
Dimensionality Reduction: PCA played a vital role in reducing the complexity of the data while maintaining predictive power.
Model Tuning: Using GridSearchCV to tune the hyperparameters allowed us to fine-tune the model, resulting in an optimal performance.
Model Evaluation: Evaluating the model using various metrics and confusion matrix gave a complete picture of its strengths and weaknesses, ensuring that it can perform well in real-world scenarios.
Future Plans
Incorporating More Data: Larger datasets would help improve the generalizability of the model.
Trying Different Models: Testing algorithms like Random Forests or Support Vector Machines (SVM) might improve performance further.
Feature Engineering: Extracting more meaningful features from the gene expression data could boost the model’s accuracy.
Conclusions
Machine learning has proven to be an invaluable tool in predicting leukemia types. By using Logistic Regression combined with dimensionality reduction techniques, we were able to build a model that accurately classifies ALL and AML based on gene expression data. While challenges remain, future advancements in genomics and machine learning can further refine these models, making a significant impact on healthcare diagnostics.
Subscribe to my newsletter
Read articles from Gaurav Bhatnagar directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
