PART - 1 : EmbedAnything - Way to bootstrap the experimentation of different embedding models across different sources
 Sangeetha Venkatesan
Sangeetha Venkatesan
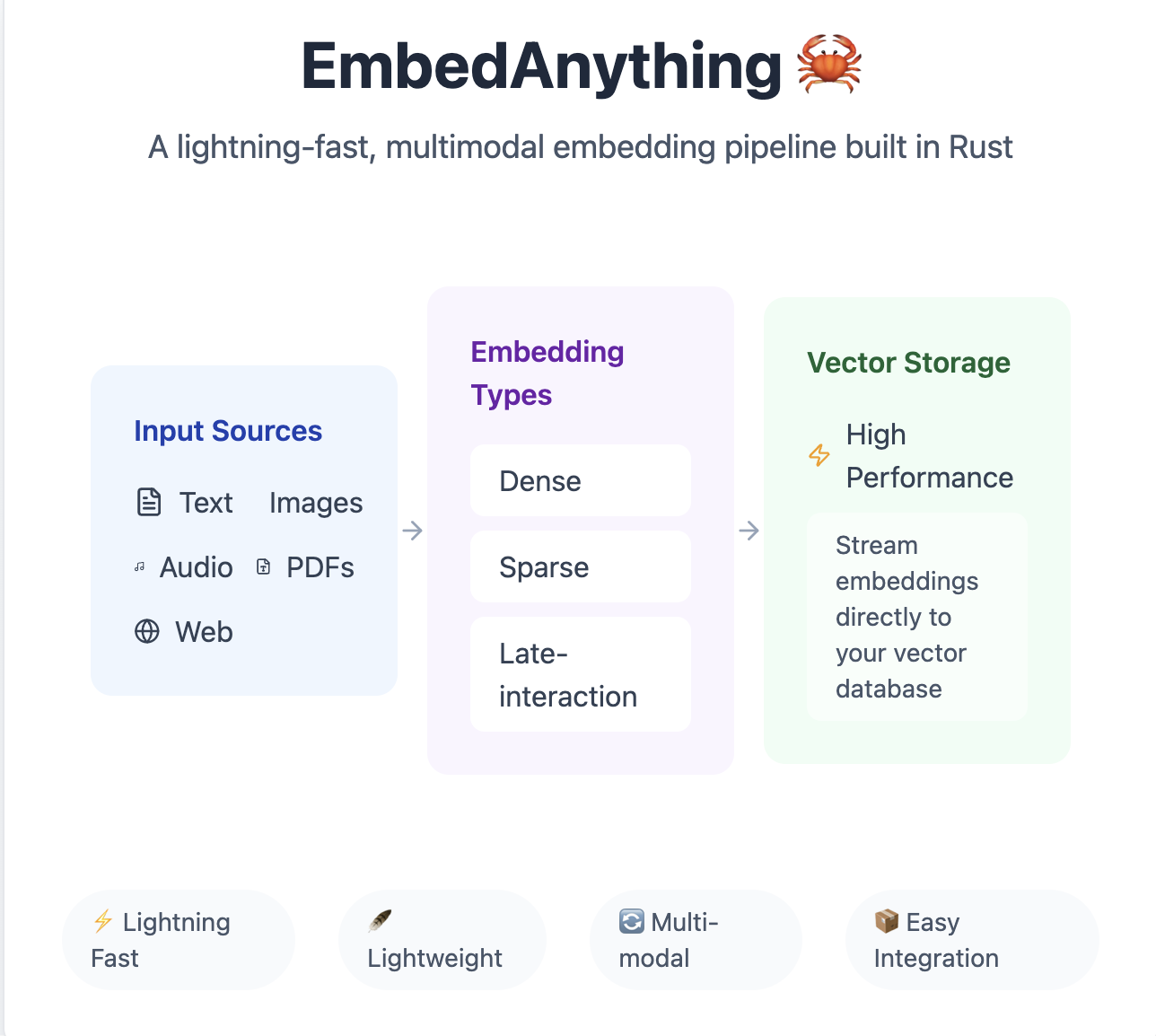
Source: https://github.com/StarlightSearch/EmbedAnything
Introduction to EmbedAnything:
To have a quick roadmap of EmbedAnything and its value proposition - https://starlight-search.com/blog/2024/12/15/embed-anything/ clearly lays out the need of inference for the embeddings on Rust backend, what’s the roadmap ahead on the model inference which helps to align where this package would be helpful in your own production or POC pipeline. It effectively exposes a python package with the function implemented in Rust through a binding interface (for building python bindings on Rust implementation).
What do you look for when you generate vector embeddings for the data? 1) Time to get the embeddings 2) Multi-modality support 3) Light-weight package (memory efficient) 4) Can handle larger amounts of data points 5) Efficient vector streaming [Kind of lazy loading to not exhaust the entire memory of loading dataset and computing all data points at once in memory] 6) Good for benchmarking multiple models at reduced latency 7) Support good range of open-source models serving. 8) Support for GPU/CPU[ based loading. 9) Support for models with contextual embeddings that takes query alongside documents into scope when calculating embeddings [Giving stronger signals of which parts of documents are important] 10) A good performance-speed-size tradeoff in execution.
Key takeaways:
The blog covers overall features embedAnything library covers with explanation of each component and how it can be leveraged to speed up the embedding workload process. Starting with the highlights on the features it supports from latest release - https://starlight-search.com/blog/2025/01/01/modernBERT/ to the complete library structure so contributors can explore avenues of scopes they can add value on back to the open-source project. The readers can get to understand different components like Chunkers, File-processors etc, Library structure, discussion on modality, Vector database adapters, Streaming, Re-rankers. Its a comprehensive overview giving an idea of all the components one by one along with the gist of how it can be leveraged to your own use-case.
Finally, we will see list of use-cases around the library and the scope we will be covering in the next-blog series in EmbedAnything.
EmbedAnything is written in Rust and it utilizes HuggingFace Candle Library (https://github.com/huggingface/candle) to serve highly performant models in the hub in faster. For generation models we use Tokens per second, Time for first token as a factor of efficiency. In the case of embedding models - efficiency counts in with faster embedding time, support of vector streaming accounting for larger scale of data in Knowledge Base.
List of Components in the package:
- Chunkers [Cumulative, Statistical]
This is used as a part of Embedding files and in directory, where each file goes through these process for faster chunking, embedding. Main focus here was to perform optimized memory efficient operations.
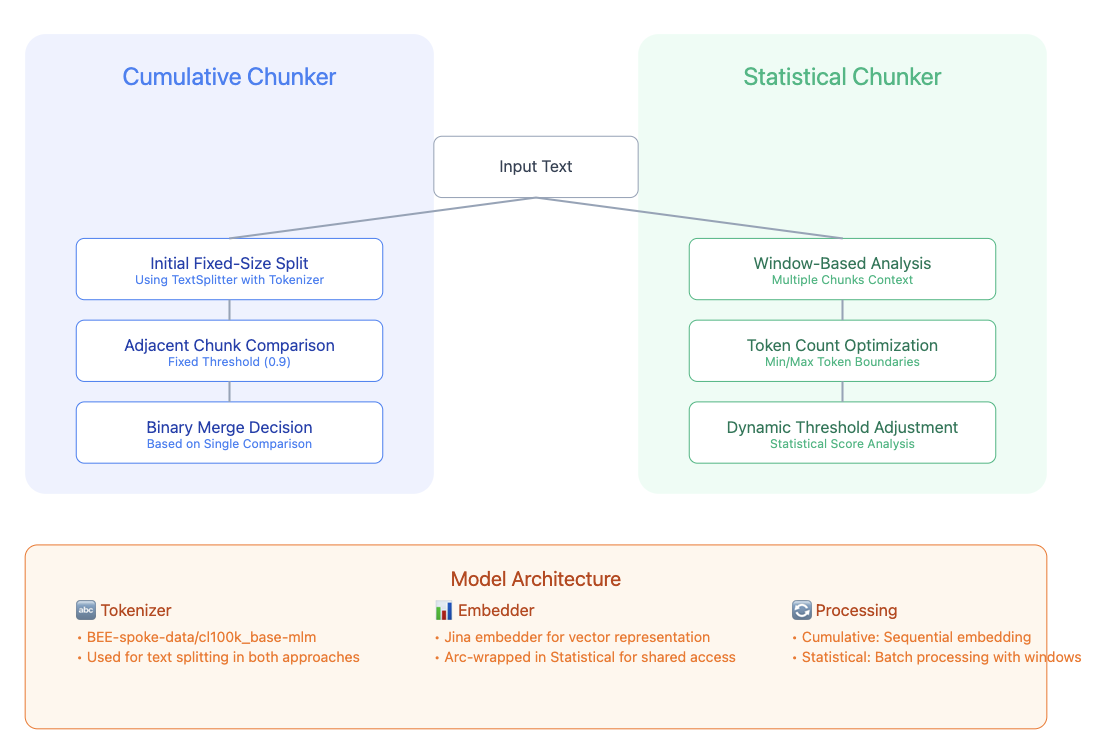
Embeddings [Local, Cloud]
File Processor [Audio, PDF, Docx, HTML, Text, Weblinks, Markdown]
Vision and other set of Retriever Models [ColPali, CLIP, ColBert, Splade]
Re-ranker
Highlights on the recent release of EmbedAnything framework:
Support for Late Interaction Models:
Enhance similarity and search process for RAG queries & Passage - Scalable and Fast!
ONNX Version of ColBert [a late interaction architecture that independently encodes the query and the document using BERT as the base model and employs a cheap yet powerful interaction step that models their fine-grained similarity] and ColPali is added as a retriever models to speed up the RAG process. I see this is much useful in adding multimodality or enhance the search performance. In production ingestion process, in Azure AI Search, https://techcommunity.microsoft.com/blog/azure-ai-services-blog/introduction-to-ocr-free-vision-rag-using-colpali-for-complex-documents/4276357 → Considering, inclusion of ColPali retriever(ColBert style) PyTorch model loading especially python wrapper is a heavy process in the ingestion and query inference, having included ONNX version of these models in ingestion is definitely a boost in production use cases.
ColPali and ColBert Quantized versions:
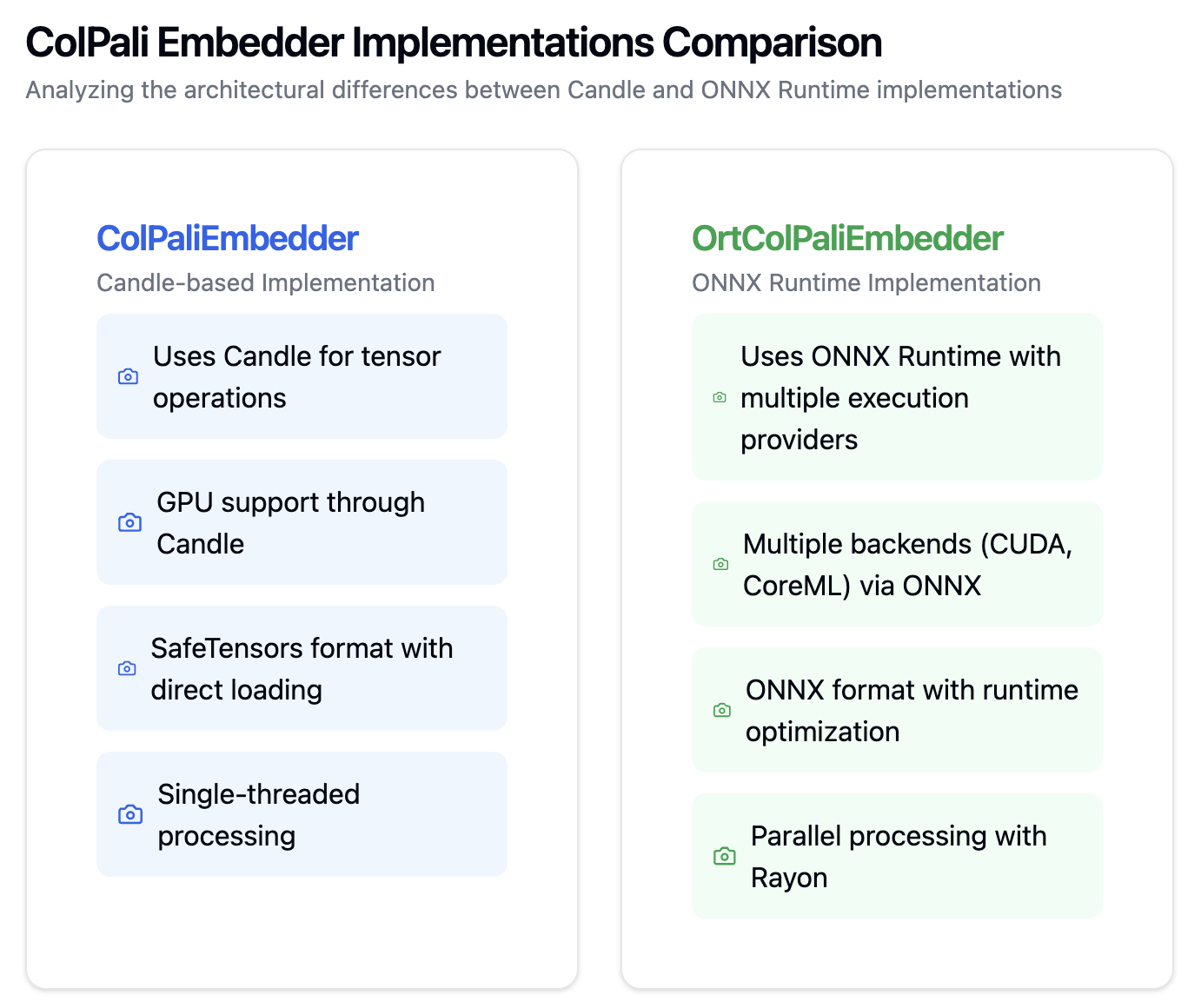
Handling of Images:
The package has support for ONNX models, Dense, Sparse Representations, Colpali and other multimodal embeddings from CLIP, JINA. Key performance benefit comes from the Rust implementation of Parallel iteration, ONNX optimization for inference, making use of the benefits on compute intensive parts.
Performing a multimodal image and query search: This is to see the overall querying and retrieval time efficiency between loading models through EmbedAnything (openai/clip-vit-base-patch16) vs the Cohere embed multimodal utility. It depends on the dimensions data points we work with, no of images etc. This is just to see the overall loading and computation aspect and not model performance comparison. This is to see how much these optimizations augments the whole pipeline on a larger scale of images and documents.
Considering I have 5 images of Japanese food in the image directory to search across which has Ramen.
Query: ["Feeling like having a bowl of hot Ramen"]
The possibilities of optimizations and gains from models through these frameworks makes the overall model loading, embedding and querying faster. The package looks well orchestrated in experimenting with different kinds of embedding models across various data sources. For closed source cohere model - we use model='embed-english-v3.0' alongside base64 encoded string of image.

import os
import numpy as np
from PIL import Image
import embed_anything
from embed_anything import EmbedData
import time
import matplotlib.pyplot as plt
# Start the overall timer
start_time = time.time()
# Directory containing the images
image_dir = "/content/sample_data"
# Filter the list to only include .png files
image_files = [f for f in os.listdir(image_dir) if f.lower().endswith(".png")]
if not image_files:
print("No .png files found in the directory.")
else:
# Load the model
model_load_start_time = time.time()
model = embed_anything.EmbeddingModel.from_pretrained_hf(
embed_anything.WhichModel.Clip,
model_id="openai/clip-vit-base-patch16",
)
model_load_end_time = time.time()
print(f"Time taken to load the model: {model_load_end_time - model_load_start_time:.2f} seconds")
# Embed images in the directory
image_embed_start_time = time.time()
data: list[EmbedData] = embed_anything.embed_image_directory(image_dir, embeder=model)
image_embed_end_time = time.time()
print(f"Time taken to embed images: {image_embed_end_time - image_embed_start_time:.2f} seconds")
# Convert the embeddings to a numpy array
embeddings = np.array([d.embedding for d in data])
# Print dimensions of the embedded data for each image
for i, d in enumerate(data):
print(f"Embedding dimensions for {os.path.basename(d.text)}: {len(d.embedding)}")
# Embed a query
query = ["Feeling like having a bowl of hot Ramen"]
query_embed_start_time = time.time()
query_embedding = np.array(
embed_anything.embed_query(query, embeder=model)[0].embedding
)
query_embed_end_time = time.time()
print(f"Time taken to embed query: {query_embed_end_time - query_embed_start_time:.2f} seconds")
# Normalize embeddings
embeddings_norm = embeddings / np.linalg.norm(embeddings, axis=1, keepdims=True)
query_embedding_norm = query_embedding / np.linalg.norm(query_embedding)
similarities = np.dot(embeddings_norm, query_embedding_norm)
# Find the index of the most similar embedding
max_index = np.argmax(similarities)
# Get the most similar image path and similarity score
most_similar_image_path = data[max_index].text
similarity_score = similarities[max_index]
print(f"Most similar image: {os.path.basename(most_similar_image_path)}")
print(f"Similarity score: {similarity_score:.2f}")
# Display the most similar image inline using matplotlib
image = Image.open(most_similar_image_path)
plt.imshow(image)
plt.axis("off") # Hide axis for better display
plt.title(f"Most Similar Image: {os.path.basename(most_similar_image_path)}\nSimilarity: {similarity_score:.2f}")
plt.show()
# End the overall timer
end_time = time.time()
print(f"Total time taken: {end_time - start_time:.2f} seconds")
Support of ModernBERT & ColBERT style models:
ModernBERT's architecture offers 3-6x faster inference than traditional BERT models. Having a different paradigm of processing in encoder models is needed to transform the ingestion pipeline. Also this offers higher token window when compared to 512 tokens encoder only models. Applying the concept of ModernBERT to Embeddings is what the https://huggingface.co/nomic-ai/modernbert-embed-base Nomic AI brought to the table. Embedding models - major advantage comes into the retrieval time latency - with lesser dimensions using “Matryoshka Representation Learning” - significant memory savings (embeddings are sorted in order of importance) - Pareto improvement over BERT - that ModernBERT improves both speed and accuracy compared to BERT (or similar models), without sacrificing one to improve the other. In decoder land - these improvements in reasoning become evolutionary. Hence making both speed and retrieval efficiency go hand in hand is important.

Transformer for RAG: ModernBERT [ONNX Version - Embedding Model] - Bringing the advancements of ModernBERT to embeddings
ModernBERTBase - nomic-ai/modernbert-embed-base
ModernBERTLarge - nomic-ai/modernbert-embed-large
import os
from pathlib import Path
import time
from rich.progress import Progress, SpinnerColumn, TimeElapsedColumn
from rich.console import Console
from rich.table import Table
from rich.panel import Panel
from embed_anything import EmbeddingModel, WhichModel, ONNXModel, Dtype
# Initialize Rich console
console = Console()
# Clone repository if it doesn't exist
if not os.path.exists("ProcessingTestFiles"):
console.print("[cyan]Cloning repository...[/cyan]")
os.system("git clone https://github.com/SangeethaVenkatesan/ProcessingTestFiles.git")
console.print("[green]Repository cloned successfully![/green]")
# Set directory path to the test_files folder in the cloned repo
directory = Path("ProcessingTestFiles/test_files")
console.print(Panel(f"[bold green]Using directory:[/bold green] {directory}", title="Setup"))
# Initialize EmbedAnything models
models = [
(EmbeddingModel.from_pretrained_onnx(WhichModel.Bert, ONNXModel.ModernBERTBase, dtype=Dtype.Q4F16), "NomicAI-ModernBERT-Base")
]
# Process all files in the directory
files = list(directory.glob("*.*")) # Get all files regardless of extension
console.print(f"\n[bold cyan]Found {len(files)} files to process...[/bold cyan]")
# Prepare embedding results
embedding_results = []
# Embedding progress
with Progress(
SpinnerColumn(),
*Progress.get_default_columns(),
TimeElapsedColumn(),
console=console
) as progress:
embed_task = progress.add_task("[green]Embedding Files with EA Base and Large...", total=len(files) * len(models))
for model, model_name in models:
for file in files:
try:
# Start timing
start_time = time.time()
# Embed file using the model
embedding_list = embed_anything.embed_file(str(file.absolute()), embedder=model)
# Parse embeddings and extract dimensions
if embedding_list and isinstance(embedding_list, list):
embedding_dim = len(embedding_list[0].embedding) if hasattr(embedding_list[0], "embedding") else "N/A"
else:
embedding_dim = "N/A"
# Calculate embedding time
elapsed_time = time.time() - start_time
# Collect results
embedding_results.append({
"file_name": file.name,
"model_name": model_name,
"file_size": file.stat().st_size / 1024, # File size in KB
"embedding_time": elapsed_time,
"embedding_dim": embedding_dim,
})
progress.advance(embed_task)
except Exception as e:
console.print(f"[bold red]Error embedding file {file} with {model_name}: {e}[/bold red]")
# Create summary table
table = Table(title="Embedding Results")
table.add_column("File Name", style="magenta", justify="left")
table.add_column("Model", style="yellow", justify="left")
table.add_column("File Size (KB)", style="cyan", justify="right")
table.add_column("Embedding Time (s)", style="green", justify="right")
table.add_column("Embedding Dimension", style="blue", justify="right")
# Populate table
total_size = 0
total_time = 0
for result in embedding_results:
total_size += result["file_size"]
total_time += result["embedding_time"]
table.add_row(
result["file_name"],
result["model_name"],
f"{result['file_size']:.2f}",
f"{result['embedding_time']:.2f}",
str(result["embedding_dim"])
)
# Add total row
table.add_row(
"[bold]Total[/bold]",
"[bold]--[/bold]",
f"[bold]{total_size:.2f}[/bold]",
f"[bold]{total_time:.2f}[/bold]",
"[bold]--[/bold]"
)
console.print("\n")
console.print(table)
Above can be used as a template to put different models embedAnything framework supports, load the model, perform the calculation of document embeddings, dimensions, latency.
The Embeddings from the functions in EmbedAnything follows the structure of EmbedData which consists of following components with the support of interpreting in dense and multi-vector representations. This forms the important structure to note on handling the embeddings and performing further manipulations with it alongside feeding metadata into the generation model for the reasoning task.
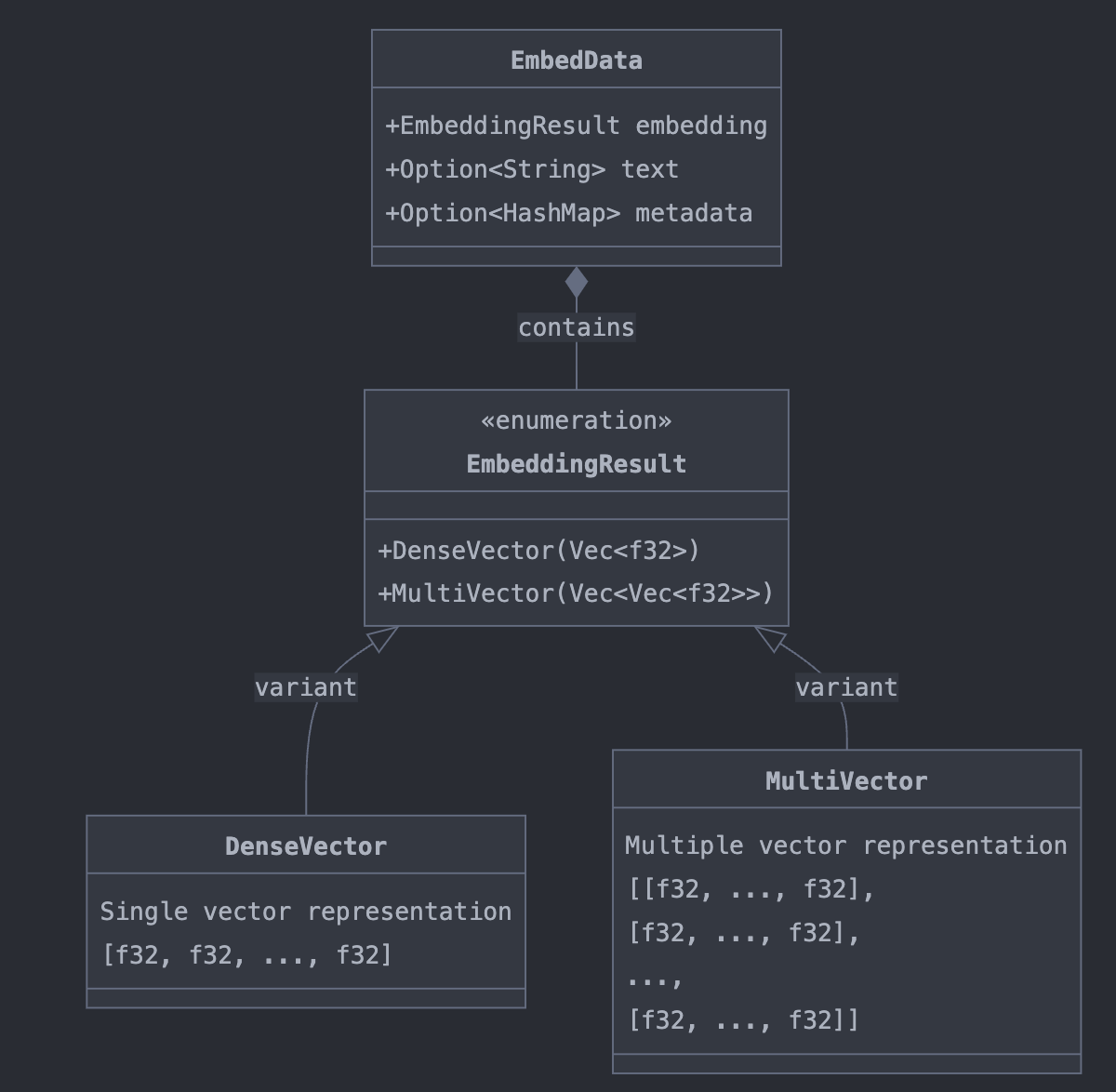
These can be chained together down the tasks of retrieval and generation. In Azure AI Search - average search time for a vector Search is 0.3 seconds and the embeddings creation of index of documents + query depends on the model used - If the models can be served through the RUST framework - the time to index and query would be substantially reduced.
Support of COLBERT style of models for retrieval:
Instead of compressing the document into single embedding representation, Contextualized embeddings added good accuracy in the retrieval process - where we break down the document and query into token level embeddings and perform a maximum similarity across each document alongside query tokens. Below picture from Langchain explains the difference between traditional vector search and contextualized late interaction method of embedding calculation.
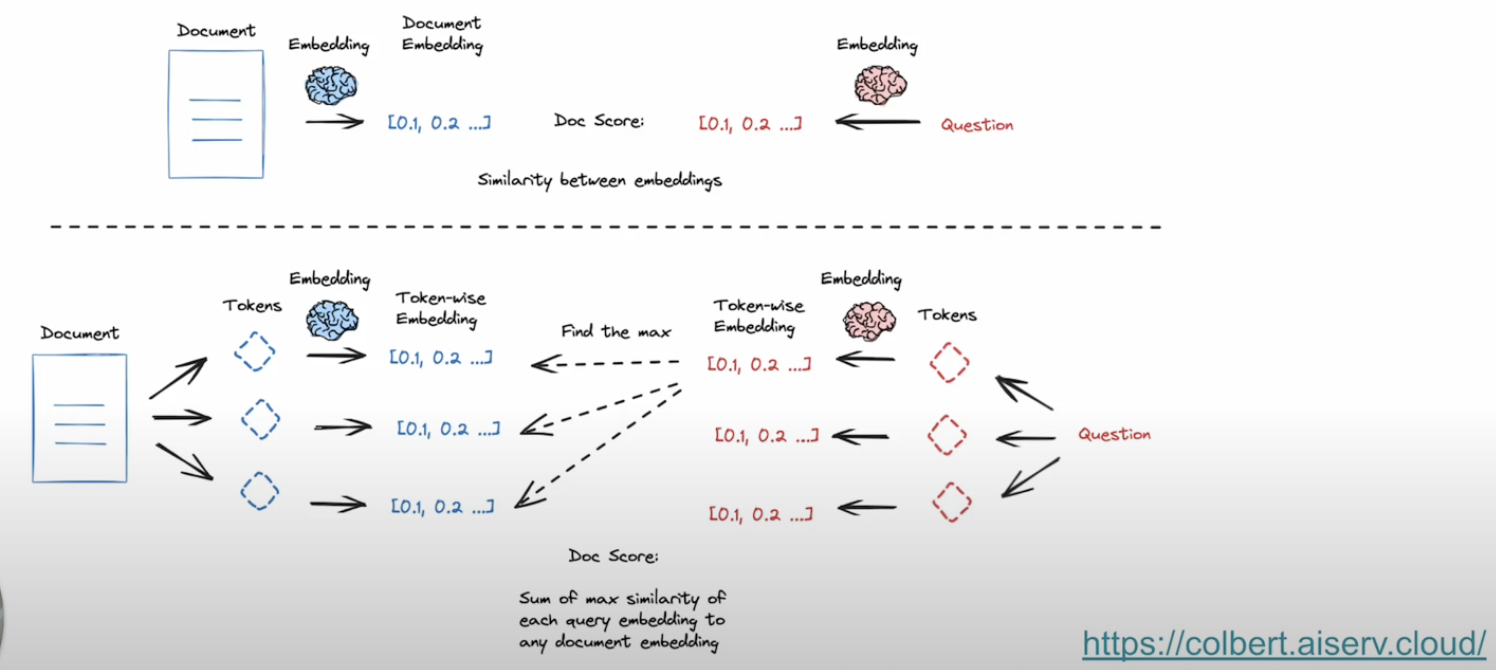
answerdotai/answerai-colbert-small-v1
jinaai/jina-colbert-v2
jinaai/jina-colbert-v1-en
Since the number of computations and dimensions handling is large when compared to single vector embedding - having a efficient backend of manipulations on larger set of documents becomes essential.
Serving LLM Models:
There are backends like VLLM, LitServe, Ollama, FastEmbed (Embedding Generation) to make the serving efficient. Specifically targeting Embedding models, that’s very crucial in RAG use cases alongside text extraction, chunking, embedding, meta-data extraction. Models in HuggingFace supported by Candle and also the adapters on the ONNX version is given in the library. Sentence transformers with the backend configuration of onnx specification does allow to get embeddings faster - but having a RUST backend has its advantages on memory handling and parallel efficiency.
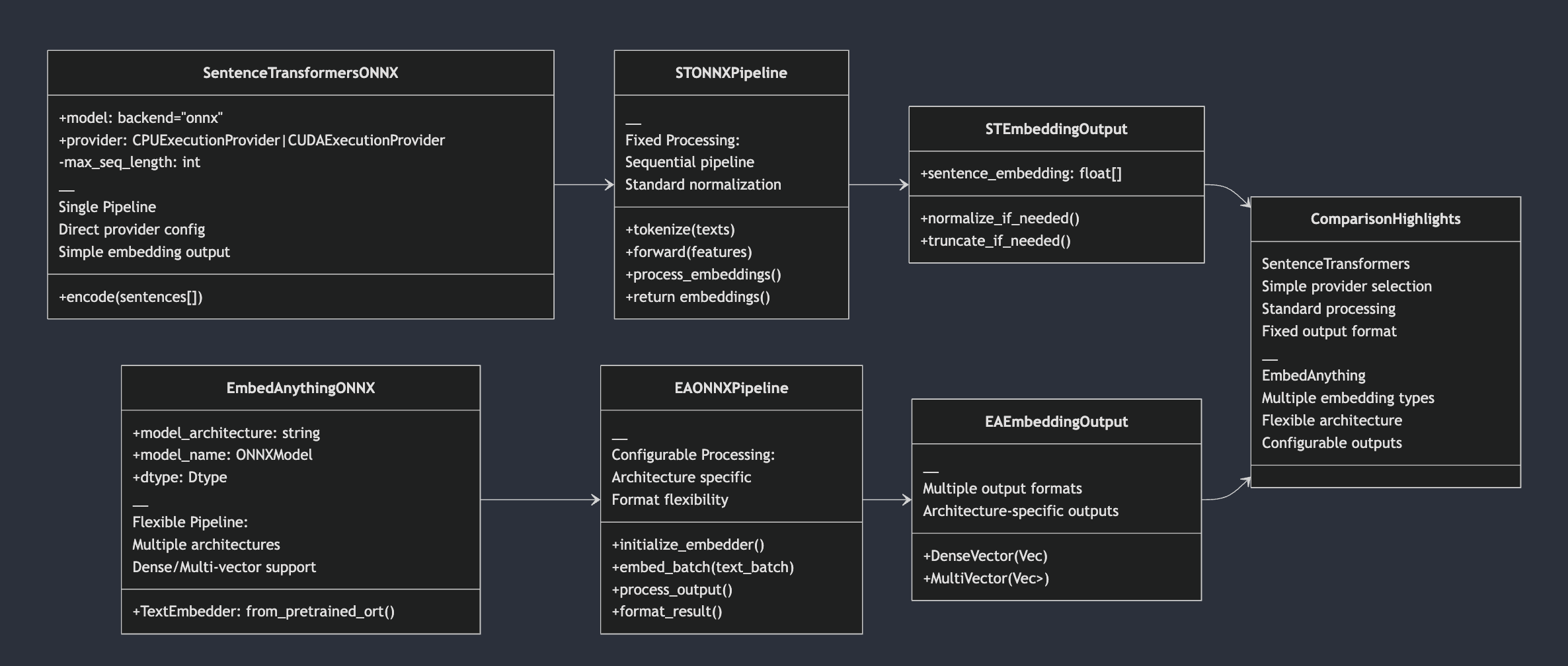
Considering embedding from EmbedAnything framework of ONNX execution vs SentenceTransformers with ONNX backend support,
SentenceTransformers
Optimization-focused pipeline
Standardized optimization levels (O1-O4)
Built-in quantization support
Single execution path
embedAnything
Flexible model architecture support
Population of text alongside embeddings, metadata - base64 encoded string
if its images
Multiple embedding types
Configurable runtime options
Diverse output formats
Impact of Candle Library for the inference of models:
[https://huggingface.co/spaces/lmz/candle-llama2]
Rust is a good candidate for these tasks when compared to python based library for its performance, memory usage (Rust ensures memory safety without needing garbage collection), concurrency. The motivation here is moving away from the Pytorch based inferencing and using Rust as a backend to boost the serving. When you want to experiment on different Embedding models from transformers library moving away from the heavy PyTorch implementations, Rust based Candle framework comes to the rescue. When targeting multiple modalities, parallel execution pipeline reduces the ingestion time since text extraction, chunking, embedding and producing vectors, streaming the vectors happens in parallel.
There is definitely a tight ecosystem between HuggingFace support of models and Candle framework, hence Candle framework alongside ONNX models are used for efficiency. GGUF Models might have a higher level edge on the efficiency when compared to Candle+ONNX.
Embedding Time Comparison:
List of Models it supports ranges from Open-source(local) and closed source(cloud)[Cohere, OpenAI] - I would say its best for running these open-source models with the powerful backend gives the quick turn-around time from ideation to production in building something large scale with greater amounts of data files.
Image Modality and Audio Modality:
When it comes to vision on images, models like ColPali are at the fore-front since it uses Vision based LLM understanding page layout ruling out OCR, text extraction and chunking. Document retrieval on a page level reduces the number of tokens sent to the vision understanding model preserving query and page layout coherence.
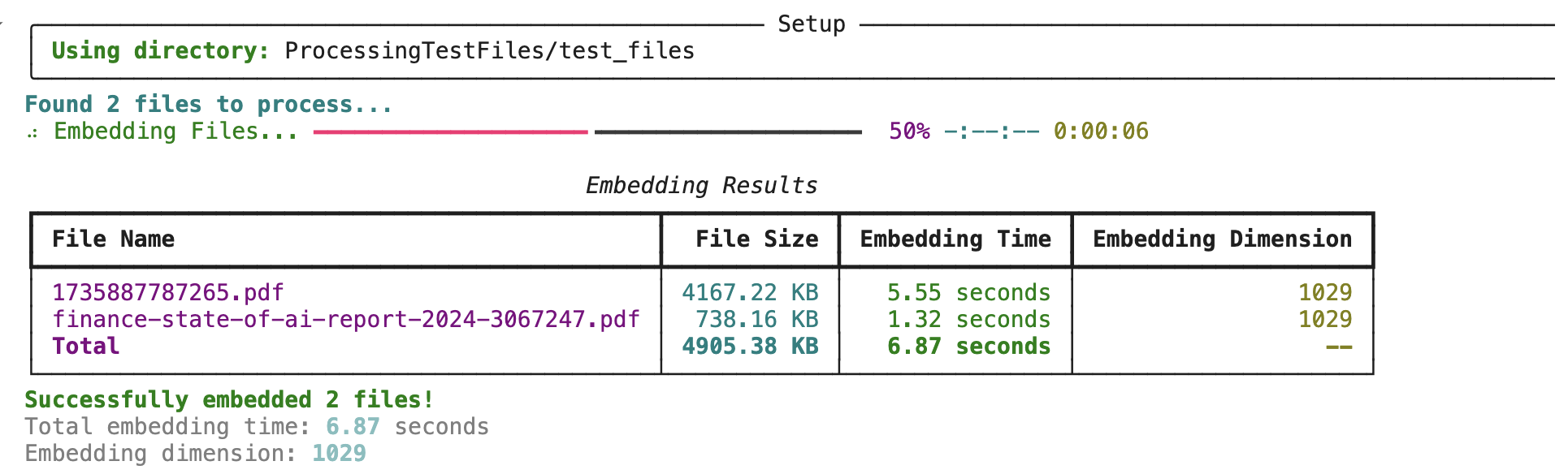
Below is the time taken using ONNX version of colPali model to get embeddings:
There are also audio encoders available in the framework to search the audio conversations which might be helpful in grouping the conversations.
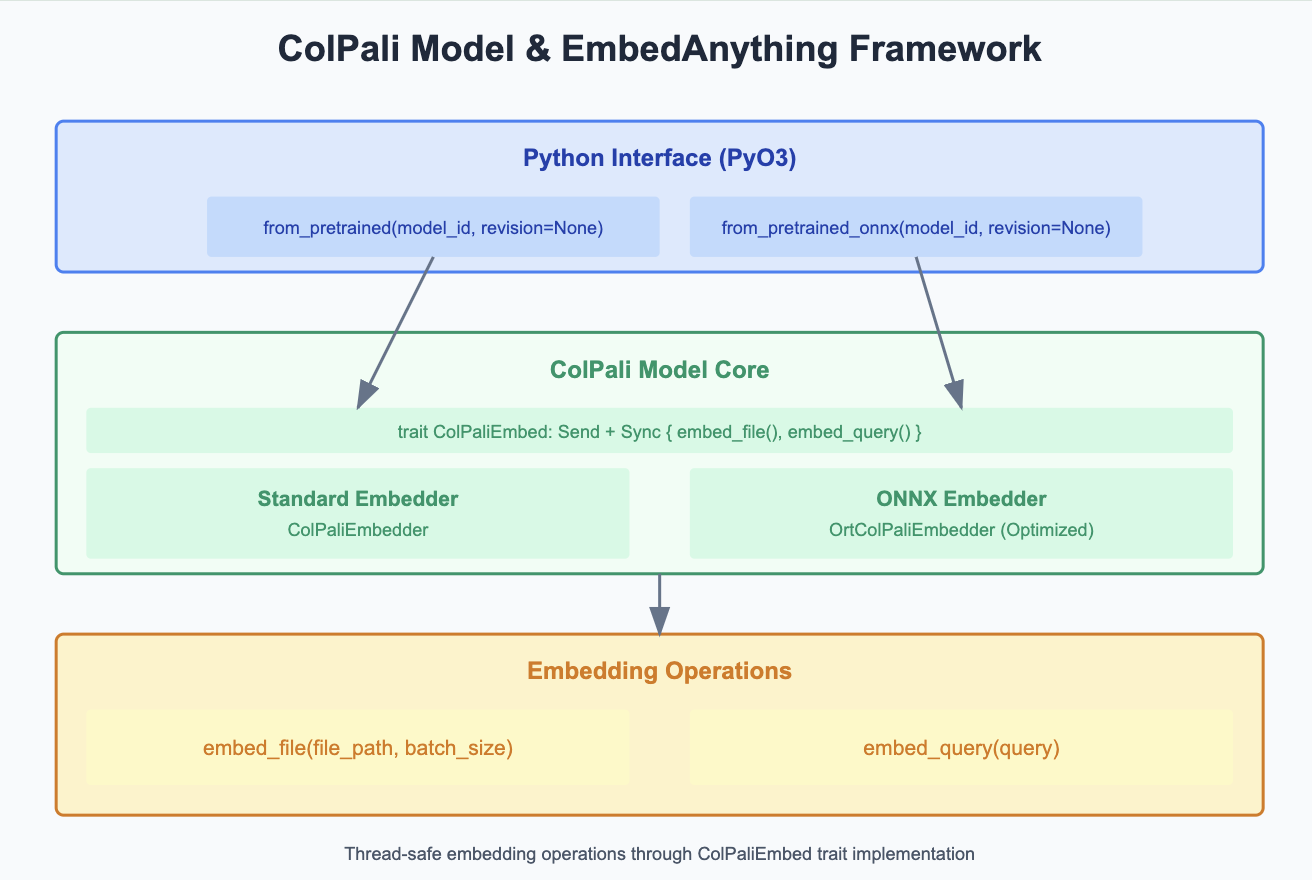
Connecting ColPali with EmbedAnything: [Inclusion of Vision based LLM]
If the document contains text as a part of Images, then these Vision LLM is very important to retrieve results. As EmbedAnything targets the latency, faster processing - Quantized version of model is introduced where we reduce the precision of model weights. Moving away from chunk free approach and lossy text extraction based methods, ColPali leverages VisionLLM to preserve the layout. Multivector embedding comes into scope here and Late interaction score is used instead of Cosine Similarity in traditional vector-based approaches. The whole ingestion pipeline can be done offline and retrieval is Page wise which is sent to Vision LLM. The Quantized version of model is served through Rust with the interface that has support for pre-trained original and onnx version. Inclusion of model always helps in reducing the space of tokens we are dealing with, the embeddings we get from these fine-tuned models helps to locate the precise space of tokens that gives correct answer thereby reducing the noise.
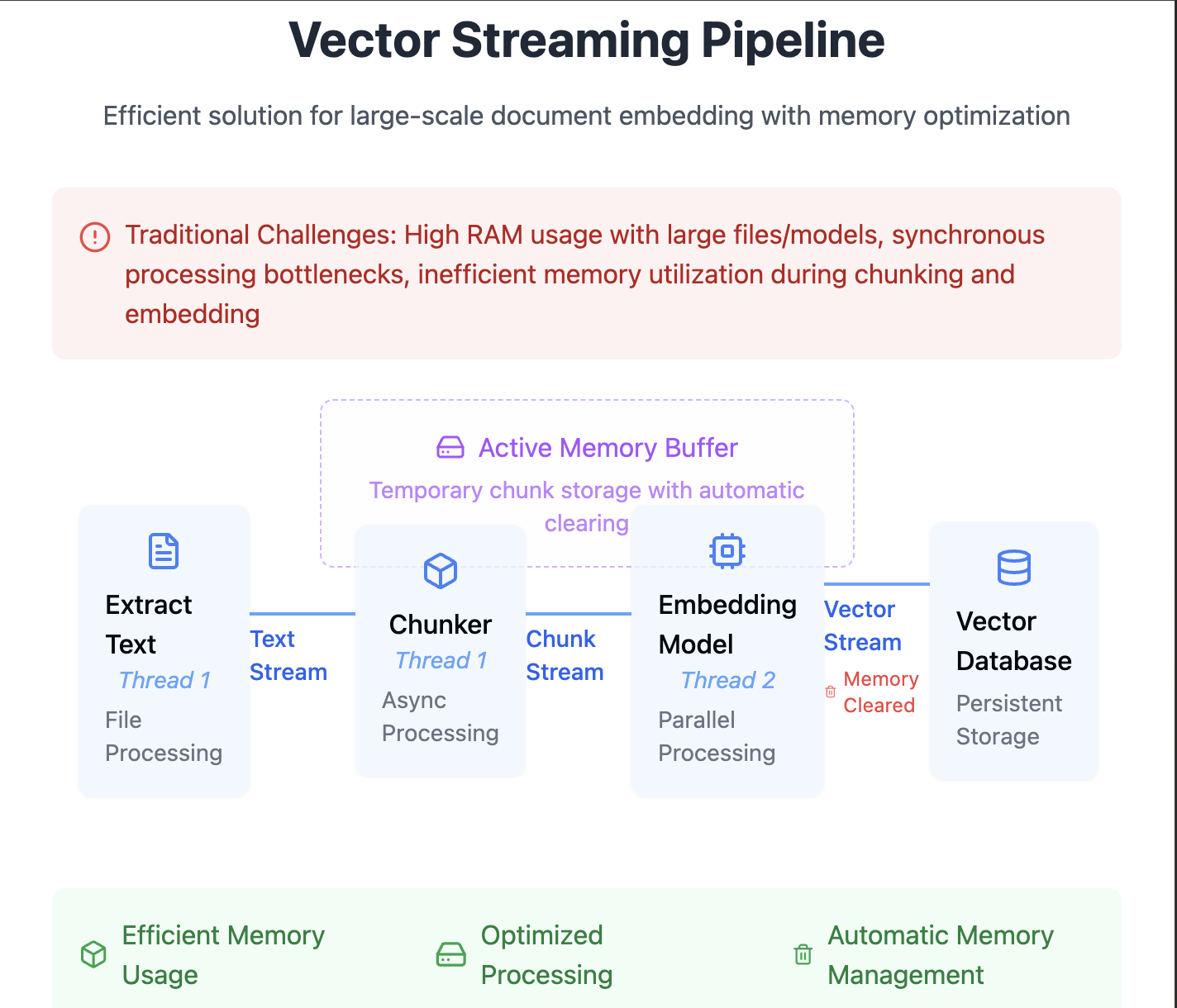
Vector Streaming:
Need of asynchronous handling here boost the performance of embedding pipeline, starting with extracting text, chunkers and exploring different embedding models. With having Rust advantage on thread safety, concurrency can be very well defined. Rust's MPSC (Multi-producer Single Consumer) help to stream the chunks into another buffer thread of embedding. Once the buffer is complete, the embedding process is done and passed back to the vector database. The process is done in memory, once moved to vector database, they are erased from memory. This is important on live scenarios like video understanding, camera feeds, malfunctioning testing, Anomaly detection. In the case of embedding background, its useful for large scale document ingestion and how it can be efficiently combined with Vectordatabase.
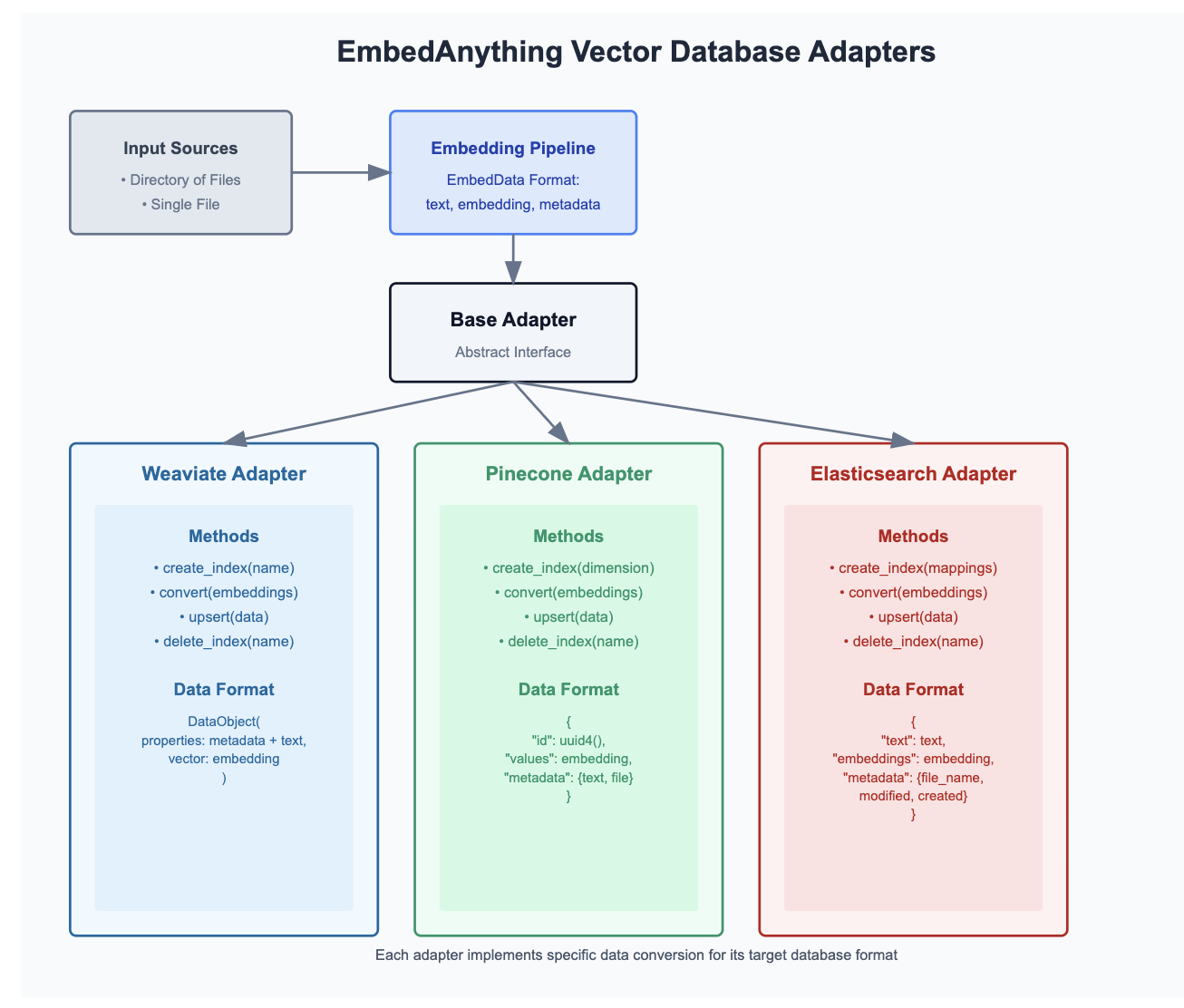
Vector database Adapters:
In order to stream files into the vector database - we can see the existing adapters EmbedAnything provides with the Vector databases and use their methods to index vectors alongside metadata as needed.
Existing list of adapters: https://github.com/StarlightSearch/EmbedAnything/tree/main/examples/adapters [Pinecone, Elastic, Weaviate]
The adapters could be customized based on the metadata the usecase needs to perform a retrieval. Inclusion of Azure AI Search would be essential to try and bootstrap the ingestion process asynchronously as Azure AI Search Index itself could allow upload around 1000 documents per API call. In one of the GitHub discussion, they have discussed on why adding more of vector database wrappers keeps it hard to be in sync, and it would become a whole lot of wrappers around. I think have this streaming efficiency Rust provides, the ingestion could be very well adapted to our own use case demanding certain aspects of metadata and methods needed to get the vector database filled in less time and memory effect.
Re-ranker:
After the retrieval, we use re-ranker models in the reduced space of documents to narrow down the choice of documents and query. Below are the list of re-rankers the library tested. For experimentation, we can compare the performance between closed source re-rankers and these on some data points serving the ONNX models through RUST embedAnything framework vs Cohere Re-ranker Models. Serving re-ranker models faster is important in real-time voice RAG applications.
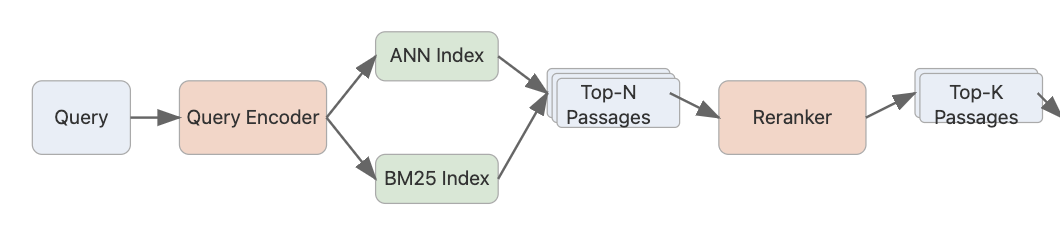
There is also notably batch embedding jobs and applying re-ranking alongside document + query token management from the closed source models like Cohere, having a framework that handles these efficiently in Rust having users make advantage of the Hugging face models.
jinaai/jina-reranker-v2-base-multilingual
jinaai/jina-reranker-v1-tiny-en
jinaai/jina-reranker-v1-turbo-en
Xenova/bge-reranker-base
Xenova/bge-reranker-large
Consider initialization of below re-ranker model from JINA AI, we can also use any fine-tuned reranker model over here for better results in the domain.
from embed_anything import Reranker, Dtype, RerankerResult, DocumentRank
from rich.console import Console
from rich.table import Table
# Initialize the console
console = Console()
# Initialize the reranker
reranker = Reranker.from_pretrained("jinaai/jina-reranker-v1-turbo-en", dtype=Dtype.F16)
# Perform reranking
query = "What is the capital of France?"
documents = ["France is a country in Europe.", "Paris is the capital of France."]
results: list[RerankerResult] = reranker.rerank([query], documents, 2)
# Display the results in a rich table
for result in results:
# Sort documents by relevance score
sorted_documents = sorted(result.documents, key=lambda x: x.relevance_score, reverse=True)
# Create a table for displaying results
table = Table(title=f"Reranked Results for Query: '{query}'")
table.add_column("Rank", style="cyan", justify="center")
table.add_column("Document", style="magenta")
table.add_column("Relevance Score", style="green", justify="center")
# Populate the table with sorted reranked documents
for rank, doc_rank in enumerate(sorted_documents, start=1):
table.add_row(
str(rank), # Use new rank after sorting
doc_rank.document,
f"{doc_rank.relevance_score:.4f}"
)
console.print(table)

Library Structure:
Hybrid Architecture - With having Python Interface, it connects to Rust bindings. Rust handles performance-critical tasks like querying and embedding. Python serves as the user-facing API, allowing easy interaction while delegating heavy computations to the Rust backend.
For those wanted to contribute to the repository, look for #[pyfunction] attribute indicates that this function is meant to be exposed to Python. On a highlevel - contributions can vary from Processors, Embeddings, Models, Re-rankers.
Use cases:
In Real-time audio based applications, latency parameter for voice RAG is extremely important, having a faster query embedding time for multiple users accessing the application in parallel.
Cases on tool calling where the gpt4o real time model works on RAG use case of embedding the queries from the user. The latency here is important to get the information out.
With the support of HTML parsing and embedding - scraping and loading the webpages layout would become easier.
When we want to adapt the embedding layer to organization own fine-tuned models, the framework supports it where we have the flexibility to increase the precision of retrieval specific to our domain.
Subscribe to my newsletter
Read articles from Sangeetha Venkatesan directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
