Article Review: Deep learning image enhancement algorithms in PET CT imaging a phantom and sarcoma patient radiomic evaluation
 Aldo Yang
Aldo Yang3 min read
Objectives
- This work quantitatively evaluates two manufacturer-developed deep-learning (DL) image enhancement algorithms for PET/CT imaging: deep learning enhancement (DLE) and deep-learning time of flight (DLT, commercialized as Precision DL).
- The evaluation uses radiomic features extracted from phantom and sarcoma patient (N=20) data, comparing DL-enhanced images to both their input images and 'gold-standard' Block Sequential Regularisation Expectation Maximisation (BSREM) with Time-of-Flight (TOF) reconstructions.
- The study demonstrates that DL-enhanced images are quantitatively similar to the 'gold-standard' images they are trained to emulate, with over 80% of radiomic features showing no significant difference.
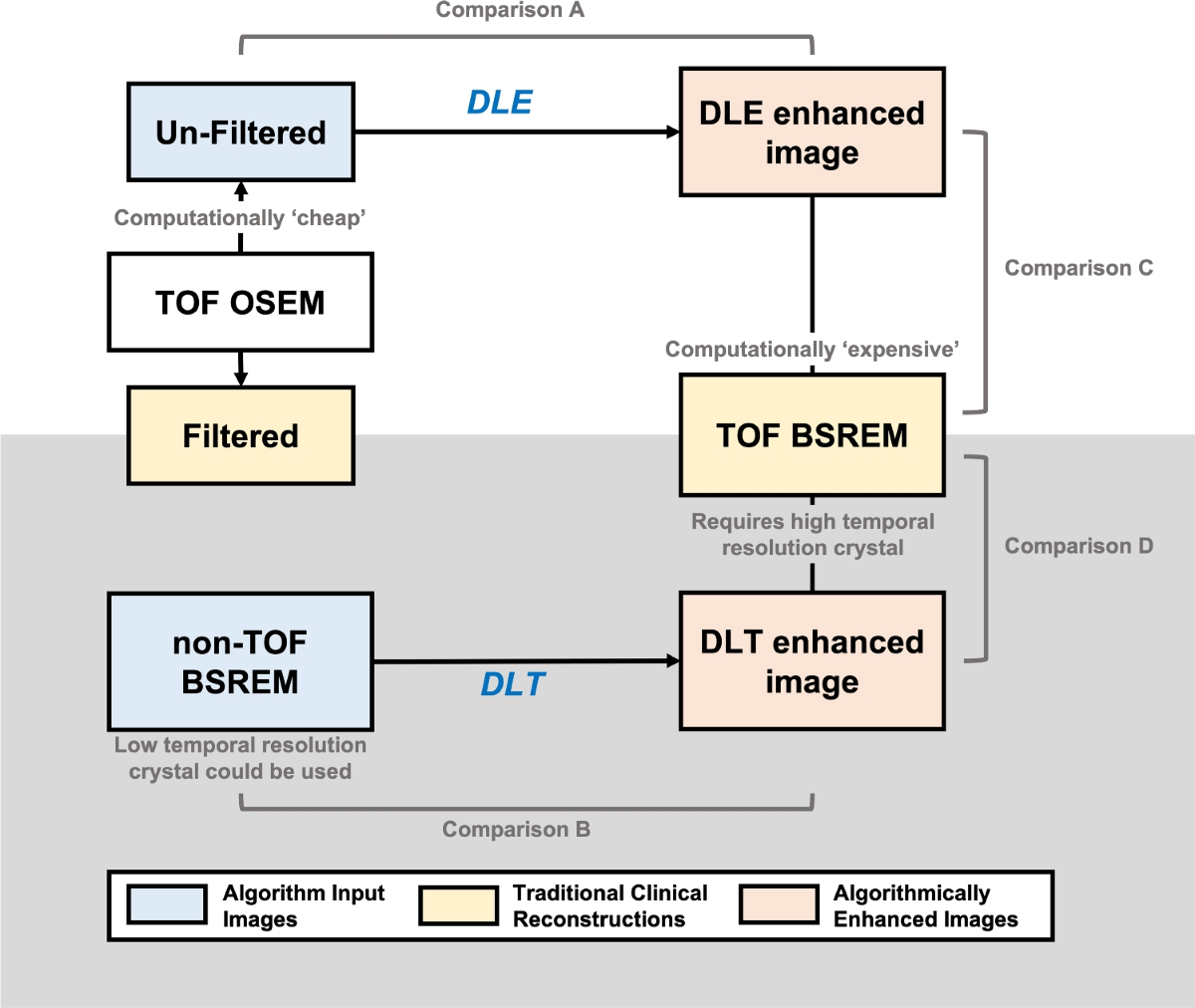
Methodology
- The study employed a retrospective sarcoma clinical [18F]FDG dataset (N=20) acquired on GE Discovery 690 or 710 PET/CT scanners, and five repeat acquisitions of a modular heterogeneous imaging phantom filled with [18F]FDG on a GE Discovery 710 PET/CT scanner.
- Six image sets were generated for each phantom acquisition and patient study: Ordered Subset Expectation Maximisation (OSEM) with filter, OSEM without filter, BSREM (non-TOF), BSREM (TOF) ('gold-standard'), OSEM without filter + DLE, and BSREM (non-TOF) + DLT.
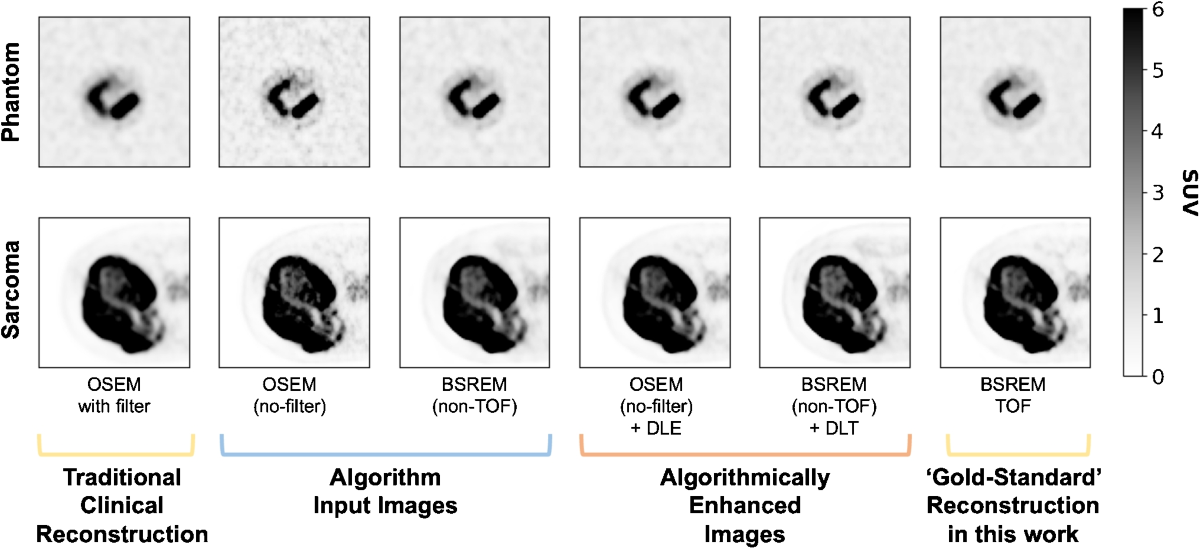
- Images were resampled to isotropic voxels (3.27 x 3.27 x 3.27 mm3). Phantom images were registered to a reference acquisition. Tumor volumes were segmented by an experienced radiologist.
- 93 International Biomarker Standardisation Initiative (IBSI) standardized radiomic features were extracted using pyradiomics.
- Statistical analysis involved paired t-tests (phantom) and t-tests on percentage differences (patient) with Bonferroni correction (pcritical = 0.0005) to compare radiomic features between image sets.
Results
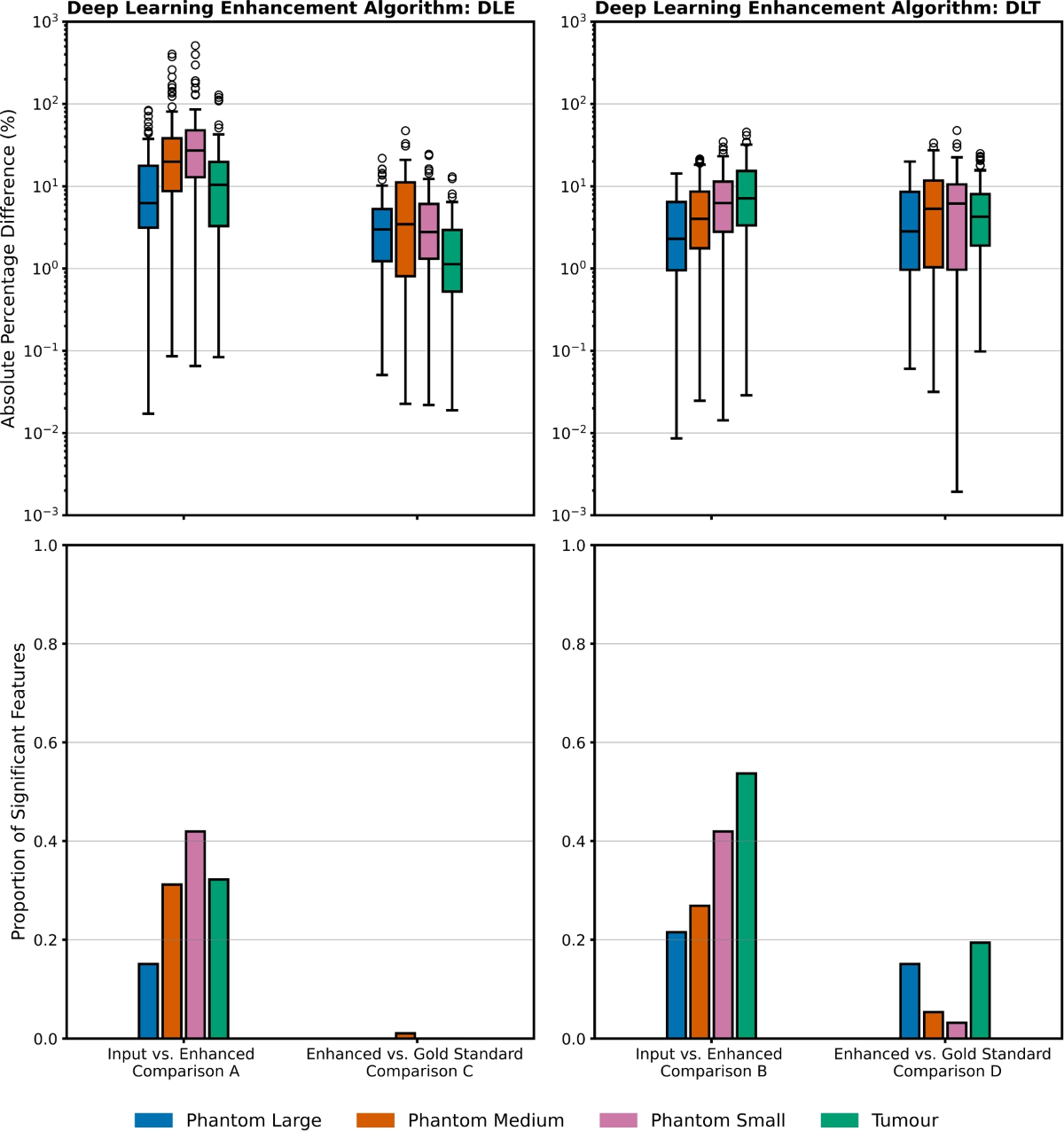
- In the phantom data, comparing DL-enhanced images to the 'gold-standard', 4.0% and 7.6% of radiomic features were significantly different (pcritical < 0.0005) for DLE and DLT, respectively.
- In the patient data, comparing DL-enhanced images to the 'gold-standard', 0.0% and 19.4% of radiomic features were significantly different for DLE and DLT, respectively.
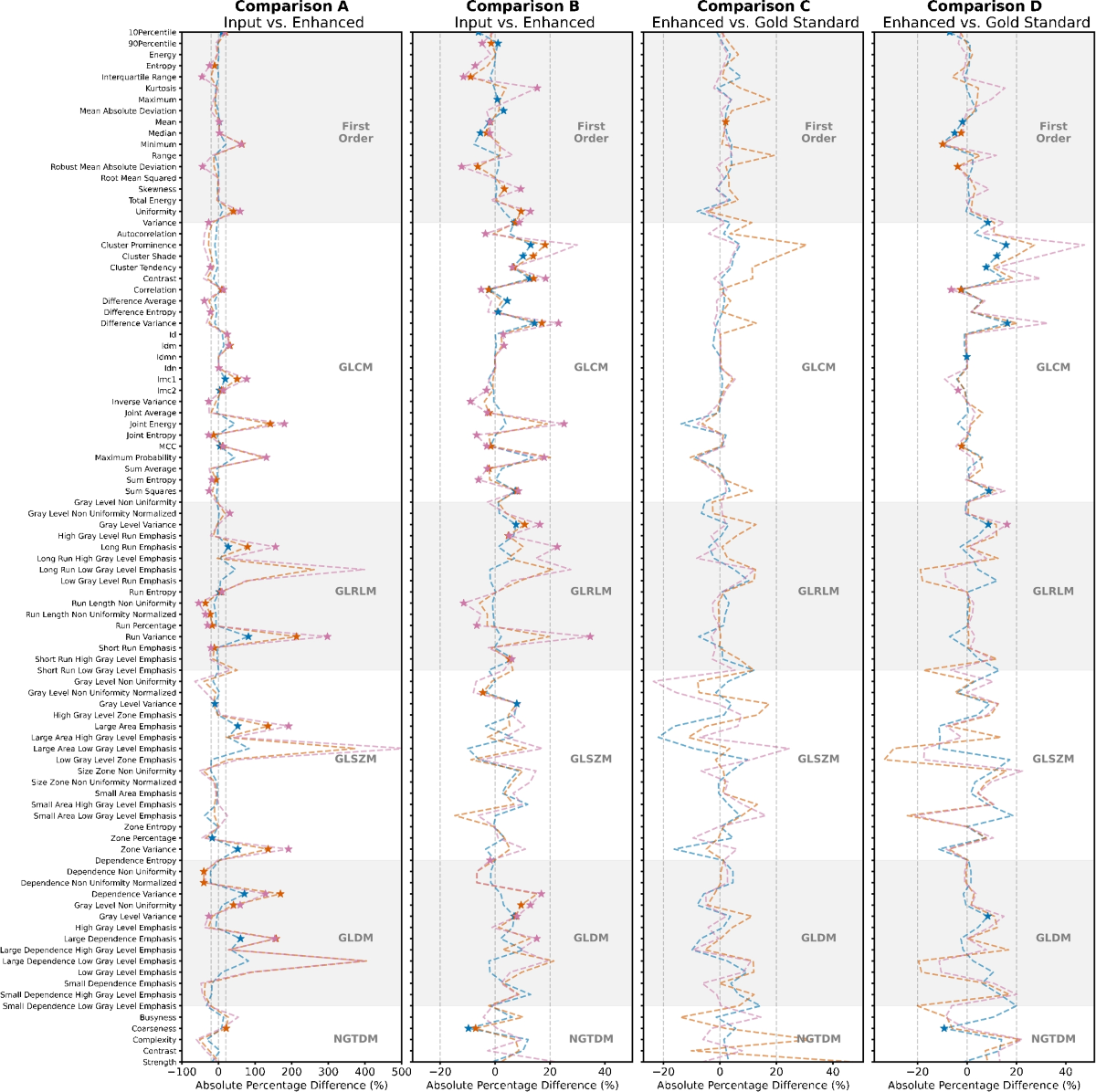
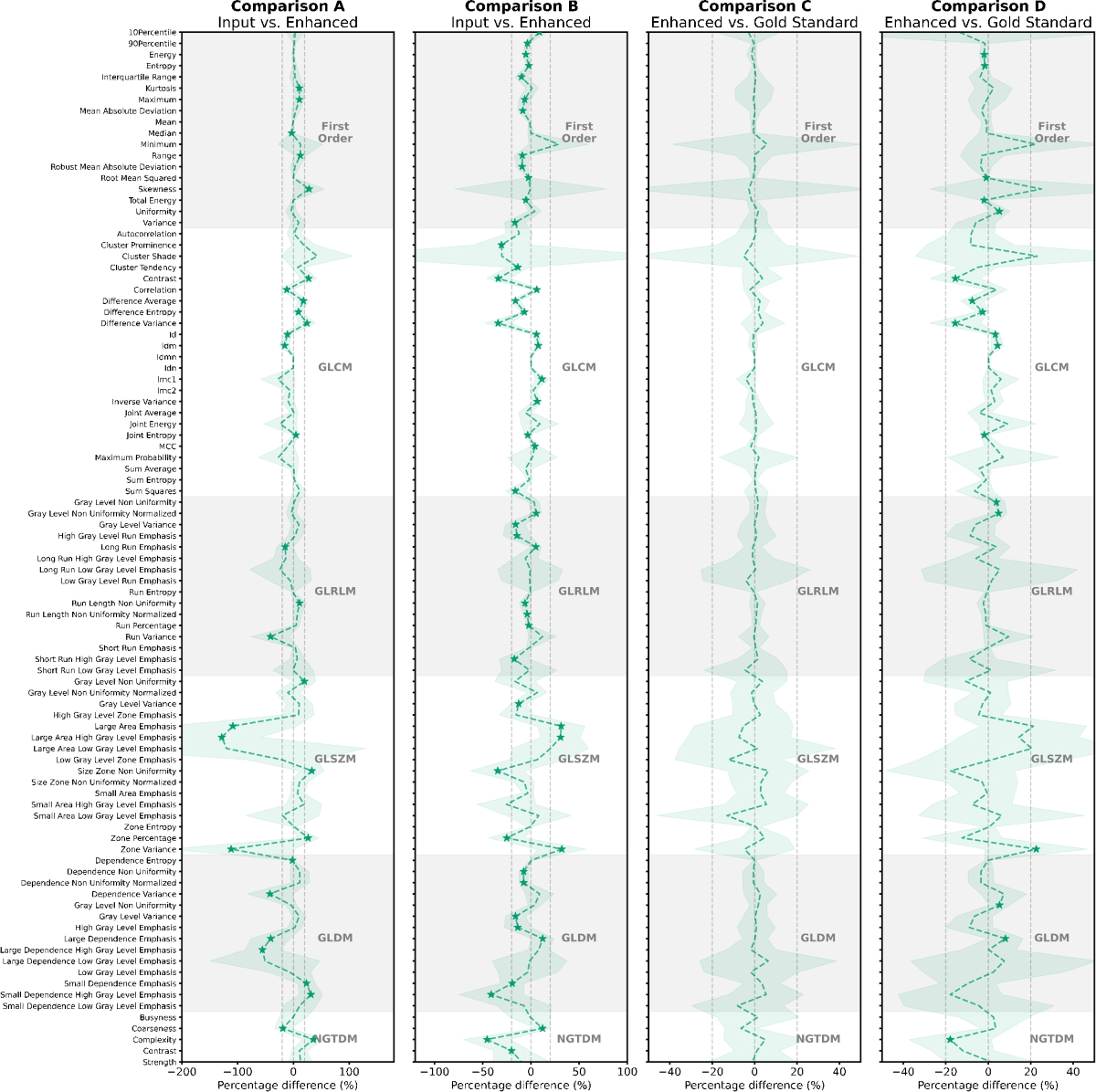
- Comparing DL-enhanced images to their input images, larger differences were observed: 29.3% and 30.3% (phantom) and 32.2% and 53.7% (patient) for DLE and DLT, respectively.
- A trend of increasing absolute percentage difference with decreasing phantom detail size was observed in most comparisons, suggesting a noise dominance in smaller details.
Discussions
- The study is well-designed and addresses an important question regarding the impact of DL image enhancement on radiomic features. However, the study is limited to a single site and a relatively small patient cohort (N=20). Expanding the study to include multi-site data and a larger, more diverse patient population would strengthen the generalizability of the findings.
- While the use of a heterogeneous phantom is a strength, the study acknowledges that no phantom can perfectly represent in-vivo activity distributions. Further investigation into the relationship between phantom characteristics and clinical results could be beneficial.
- The authors note a trend towards increased effect size with decreased detail size in the phantom data, suggesting noise dominance. A more detailed analysis of the noise characteristics of the different reconstruction methods and their impact on specific radiomic features would be valuable.
- The study uses a Bonferroni correction, which is acknowledged as conservative. Exploring less stringent correction methods could reveal additional significant differences, but the authors correctly state this would not change the main findings.
0
Subscribe to my newsletter
Read articles from Aldo Yang directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by
