Paint Your I/O Picture with Fio and Plots
 Manas Singh
Manas Singh
Fio: The chef’s knife
fio (Flexible I/O Tester) is a versatile I/O workload generator that is used to benchmark and test storage systems. Developed by Jens Axboe, fio is capable of simulating various I/O workloads, making it a valuable tool for evaluating the performance of hard drives, SSDs, and other storage solutions. fio can be configured to perform read, write, and mixed operations with different block sizes, I/O depths, and access patterns. It supports a wide range of I/O engines, including synchronous, asynchronous, and memory-mapped I/O.
Now, think of fio as a meticulously sharpened chef's knife. In the hands of a master, it can dice, slice, and julienne storage performance with unparalleled precision. It's capable of revealing the most granular details of your disks, SSDs, and network drives. But, just like a professional knife set, fio comes with a dizzying array of blades – options, in its case. This sheer versatility, while powerful, can be utterly overwhelming to the uninitiated. You're presented with a vast toolkit, and knowing which tool to use, let alone how to wield it effectively, is a challenge in itself. Prepare to navigate a sea of parameters, from block sizes and I/O engines to queue depths and latency targets, or risk simply blunting the edge of this incredibly potent instrument.
Based on the FIO documentation from FIO documentation, there are at least 152 command-line options ( as calculated by a Gemini)
Plot using the packaged script
The fio_generate_plots script is a utility that processes the log files generated by fio and creates graphical representations of the data using GNUPLOT. The script generates plots in the SVG (Scalable Vector Graphics) format, which is supported by most modern browsers and allows for resolution-independent graphs. This makes it easier to visualise and analyse the performance data collected during FIO benchmarks.
However, to create log file, certain options should be used: write_lat_log, write_bw_log, and write_iops_log. Here's a sample FIO workload configuration file:
[global]
ioengine=libaio
direct=1
bs=4k
size=1G
runtime=60
time_based
ramp_time=10s
group_reporting
numjobs=4
log_avg_msec=1000
write_bw_log=bw
write_iops_log=iops
write_lat_log=lat
[write-test]
rw=write
filename=write_test_file
[read-test]
rw=read
filename=read_test_file
To run this workload, save the above configuration to a file named fio_workload_example.ini, and then execute the following command:
fio fio_workload_example.ini
This will generate the following log files:
bw_logiops_loglat_log
These log files can then be used by the fio_generate_plots script to create the plots.
Now, to use the fio_generate_plots script to generate plots, follow these steps:
Ensure
gnuplotis installed: The script requiresgnuplotto generate graphs. You can install it using your package manager. For example, on Debian-based systems, you can use:sudo apt-get install gnuplotRun the script: Execute the script with the required parameters:
subtitle: The main title for the plots.xres(optional): The horizontal resolution of the plots.yres(optional): The vertical resolution of the plots.
Example usage:
./fio_generate_plots "Benchmark Results" 1920 1080
This will generate SVG plots with the specified resolution.
- Check the output: The script will generate SVG files in the current directory with names based on the provided subtitle and the type of data (e.g.,
My Benchmark Results-lat.svg,My Benchmark Results-iops.svg).
Make sure the script has executable permissions. If not, you can set them using:
chmod +x fio_generate_plots
On my machine, it gives an output:
> ls
Benchmark-bw.svg Benchmark-clat.svg Benchmark-iops.svg Benchmark-lat.svg Benchmark-slat.svg
Generate plots using Pandas
Now, that we have the plots as SVG, the next logical step is to automate the process and create a friendly chart. Given that there are excellent Python packages for this purpose, let us move away from gnuplot to pandas.
Here are the steps:
We read the file into a pandas data frame. Note that we are only using the first and second columns.
Collect all the data frames in a
listPlot them using matplotlib and save as SVGs
import glob
import sys
import matplotlib.pyplot as plt
import pandas as pd
def usage():
print("Usage: fio_generate_plots.py subtitle [xres yres]")
sys.exit(1)
def read_log_files(filetype):
"""
Read fio log files into a pandas dataframe.
:param filetype: string to search for filenames
:return: list of pandas dataframes
"""
logs = []
files = glob.glob(f"*_{filetype}.log") + glob.glob(f"*_{filetype}.*.log")
print(f"Found {len(files)} {filetype} files")
for file in files:
df = pd.read_csv(file, header=None, names=["time", "value", "x", "y", "z"])
df["time"] = (
pd.to_numeric(df["time"], errors="coerce") / 1000
) # Convert time to seconds
logs.append((file, df))
return logs
def plot(title, filetype, ylabel, scale, xres=1280, yres=768):
"""
Generate a plot for a given dataframe and store is as a SVG file.
:param title: Title of the plot
:param filetype: log filetype used
:param ylabel: Y axis label
:param scale: scaling factor
:param xres: X axis resolution
:param yres: Y axis resolution
:return: None
"""
logs = read_log_files(filetype)
if not logs:
print("No log files found")
sys.exit(1)
plt.figure(figsize=(xres / 100, yres / 100))
plt.title(f"{title}\n\n{ylabel}")
plt.xlabel("Time (sec)")
plt.ylabel(ylabel)
for i, (filename, df) in enumerate(logs):
depth = filename.split(".")[1]
plt.plot(
df["time"],
df["value"] / scale,
label=f"Queue depth {depth}",
linestyle="-",
marker="",
)
plt.legend(loc="best")
plt.grid(True)
plt.savefig(f"{title.replace(' ', '_')}_{filetype}.svg")
def main():
if len(sys.argv) < 2:
usage()
title = sys.argv[1]
xres = int(sys.argv[2]) if len(sys.argv) > 2 else 1280
yres = int(sys.argv[3]) if len(sys.argv) > 3 else 768
# One plot for each log type
plot(title, "lat", "Time (msec)", 1000000, xres, yres)
plot(title, "iops", "IOPS", 1, xres, yres)
plot(title, "slat", "Time (μsec)", 1000, xres, yres)
plot(title, "clat", "Time (msec)", 1000000, xres, yres)
plot(title, "bw", "Throughput (KB/s)", 1, xres, yres)
if __name__ == "__main__":
main()
Add a dash of plot
Now that we have figured out how to generate SVGs, let us take it a notch higher by creating a web app using Dash. This gives us the ability to create a dashboard and load data on demand without having to re-run scripts.
Let us re-use the read_logs_files method and replace matplotlib with plotly
Finally, wrap it in a dash app.
import glob
import pandas as pd
import plotly.graph_objects as go
from dash import Dash, html, dcc, callback, Output, Input
def read_log_files(filetype):
logs = []
files = glob.glob(f"*_{filetype}.log") + glob.glob(f"*_{filetype}.*.log")
print(f"Found {len(files)} {filetype} files")
for file in files:
df = pd.read_csv(file, header=None, names=["time", "value", "x", "y", "z"])
df["time"] = (
pd.to_numeric(df["time"], errors="raise") / 1000
) # Convert time to seconds
df.attrs["name"] = "Queue Depth" + file.split(".")[1]
logs.append(df)
return logs
app = Dash()
options = ["lat", "bw", "iops", "clat"]
app.layout = [
html.H1(children="Benchmark", style={"textAlign": "center"}),
dcc.Graph(id="graph-content"),
# Select option from the dropdown
dcc.Dropdown(options, "iops", id="dropdown-selection"),
]
@callback(Output("graph-content", "figure"), Input("dropdown-selection", "value"))
def update_graph(value):
list_of_dfs = read_log_files(value)
fig_combined = go.Figure()
for df in list_of_dfs:
for col in df.columns:
if col == "value": # Value
fig_combined.add_trace(
go.Scatter(
x=df["time"], y=df[col], mode="lines", name=df.attrs["name"]
)
)
fig_combined.update_layout(title_text="Benchmark")
return fig_combined
if __name__ == "__main__":
app.run(debug=True)
Here’s the app with a basic dropdown:
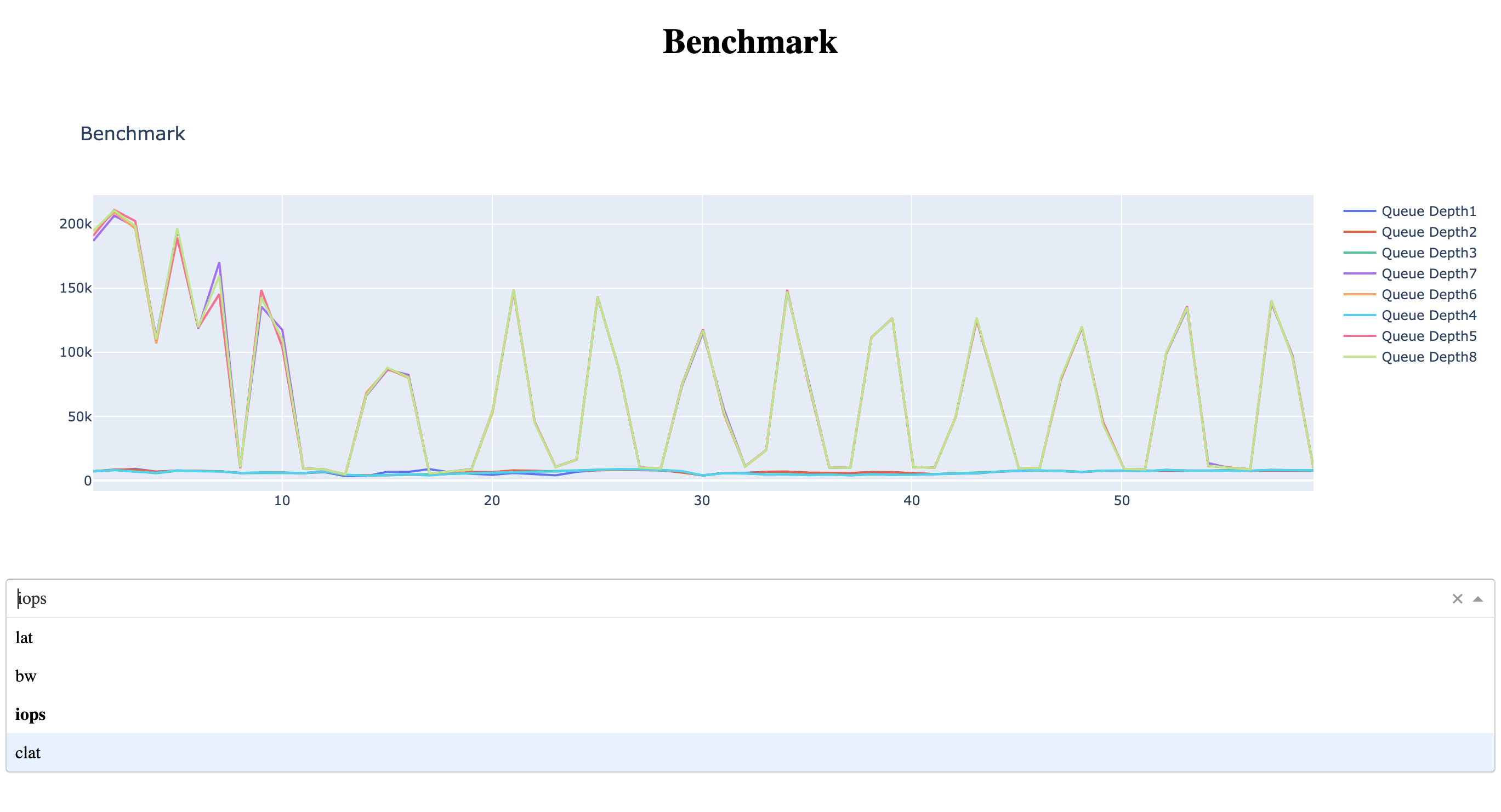
As you select the option, the app generates plot and displays them automatically.
That’s it for our fio plot app powered by Python!
Subscribe to my newsletter
Read articles from Manas Singh directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by

Manas Singh
Manas Singh
14+ Years in Enterprise Storage & Virtualization | Python Test Automation | Leading Quality Engineering at Scale