Unstructured To Structured : Using Fabric AI Functions To Extract Invoice Data From PDFs
 Sandeep Pawar
Sandeep PawarIn the long list of features announced in February's Fabric update, you might have missed a really cool new feature: Fabric AI Functions. I've written a few blogs about extracting structured data from unstructured data using AI services in Microsoft Fabric. In this blog, I will show how the Fabric AI Functions (Preview) can be used to extract structured data from PDF invoices, specifically PDFs that can be parsed as text. Fabric AI Functions are an extension of the Azure Open AI services in Fabric and provide a convenient way to use many of these AI/cognitive services seamlessly in Fabric notebooks.
Data
I will use this dataset containing real doctor bills and invoices provided by the German Research Center for Artificial Intelligence. I'm not sure if there's already published research on it; I simply searched for PDF data and found this interesting because I don't know German, making it a good use case for using LLMs. The main goal here is to demonstrate how to use LLMs and AI Functions that are natively available in Fabric.
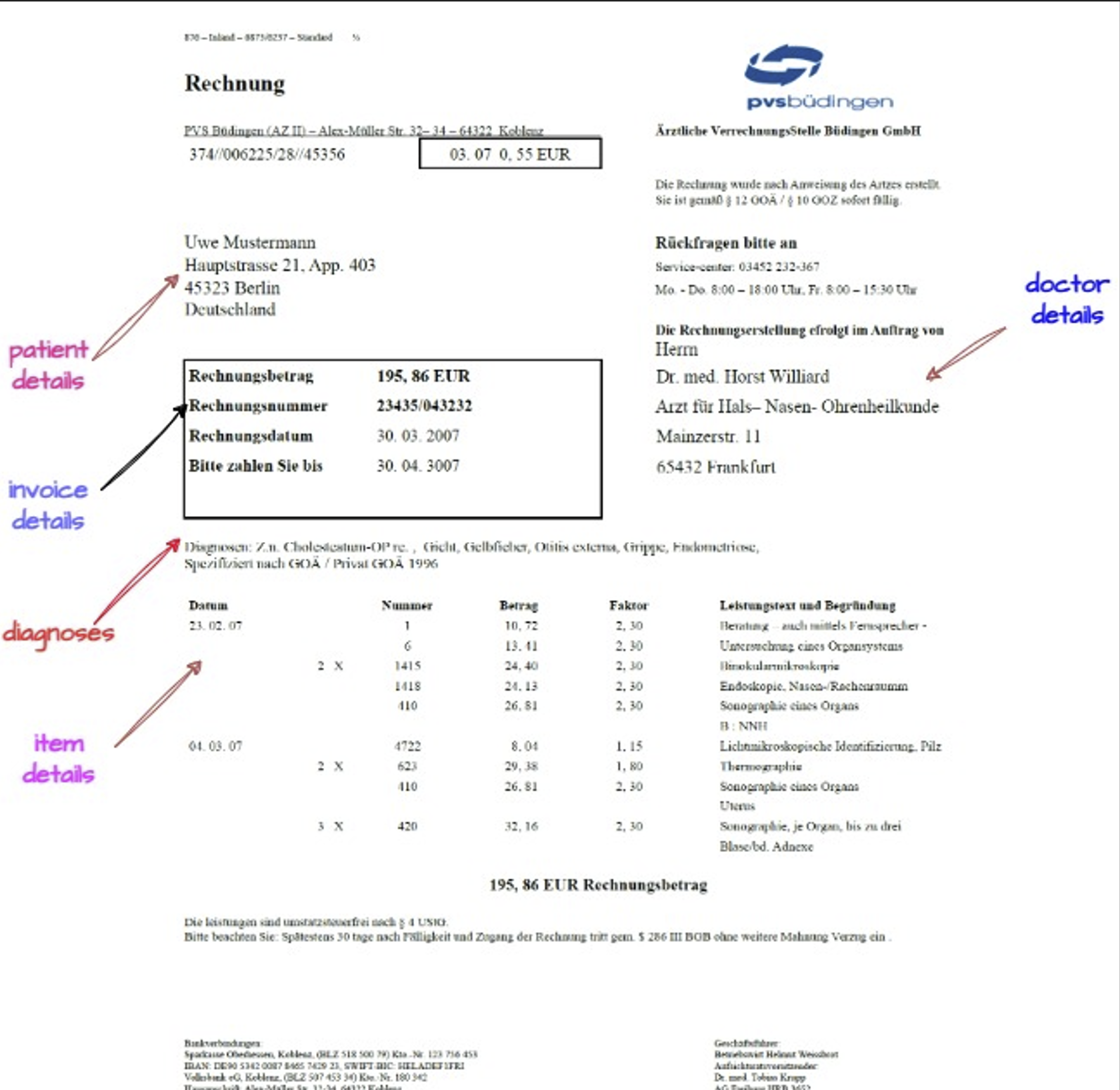
The goal here is to extract:
invoice details : invoice number, date, due date, amount
doctor details: name, specialization, address
patient details: name, address, city
diagnosis : list of diagnosis
item details table : date, number, factor, amount, description
I will approach this like any other data science project, which means having strong baselines and evaluations is important. For baselines, it's important to start with the simplest solution available and build from there. I will first use Power BI and Azure Document Intelligence for establishing baselines.
Power BI
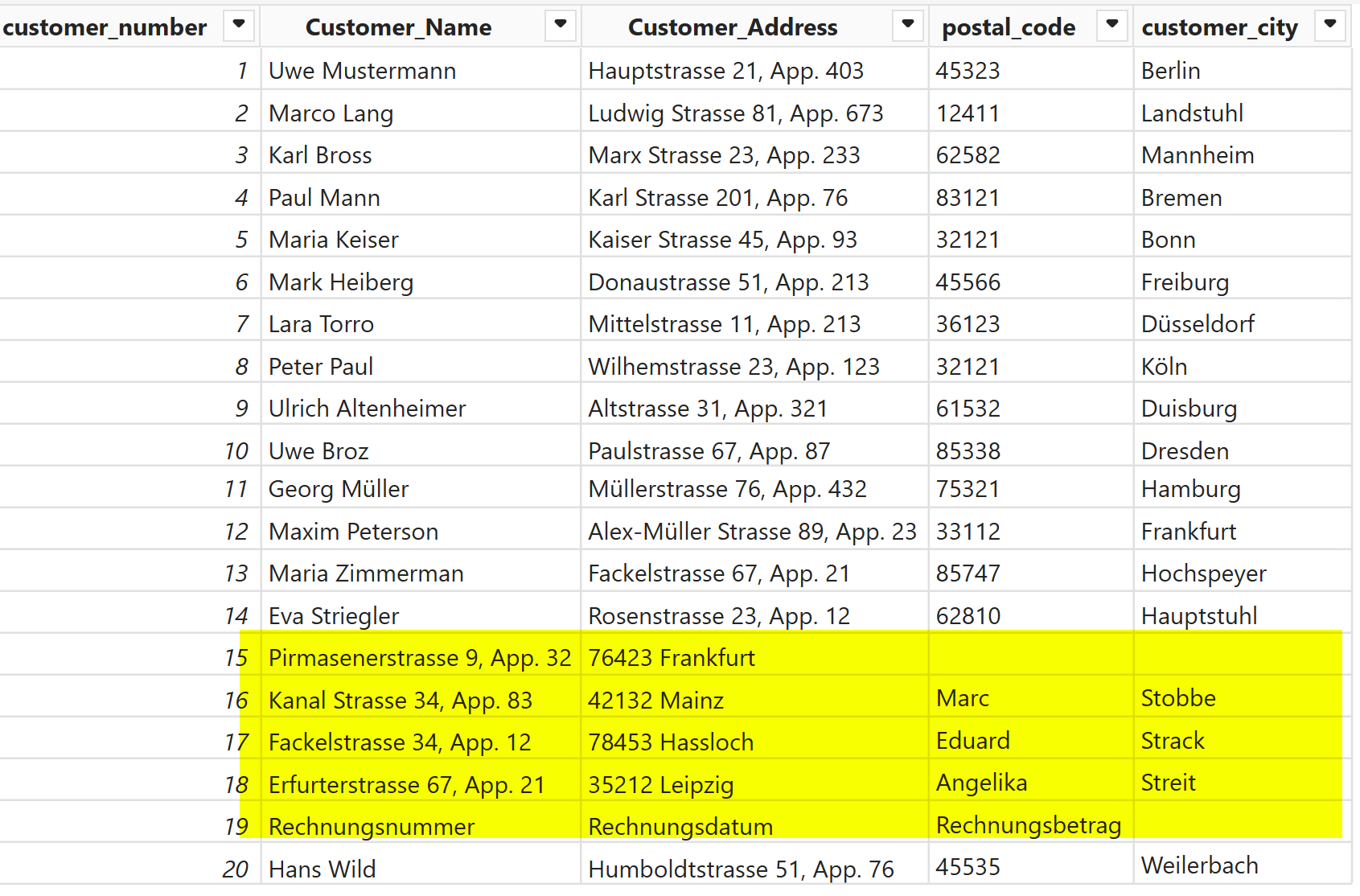
Power BI has a strong PDF parser and in the past I have been surprised by how well it works. So let’s first try that:
I imported the PDF data as a folder and did some M gymnastics to extract the patient data to start with (felt great to use M after so many months :D)
However, due to differences in layouts, not all data was extracted correctly. While this method works, it doesn't guarantee reliable results for all layouts. You can download the pbix from here and try it yourself.
Azure Document Intelligence
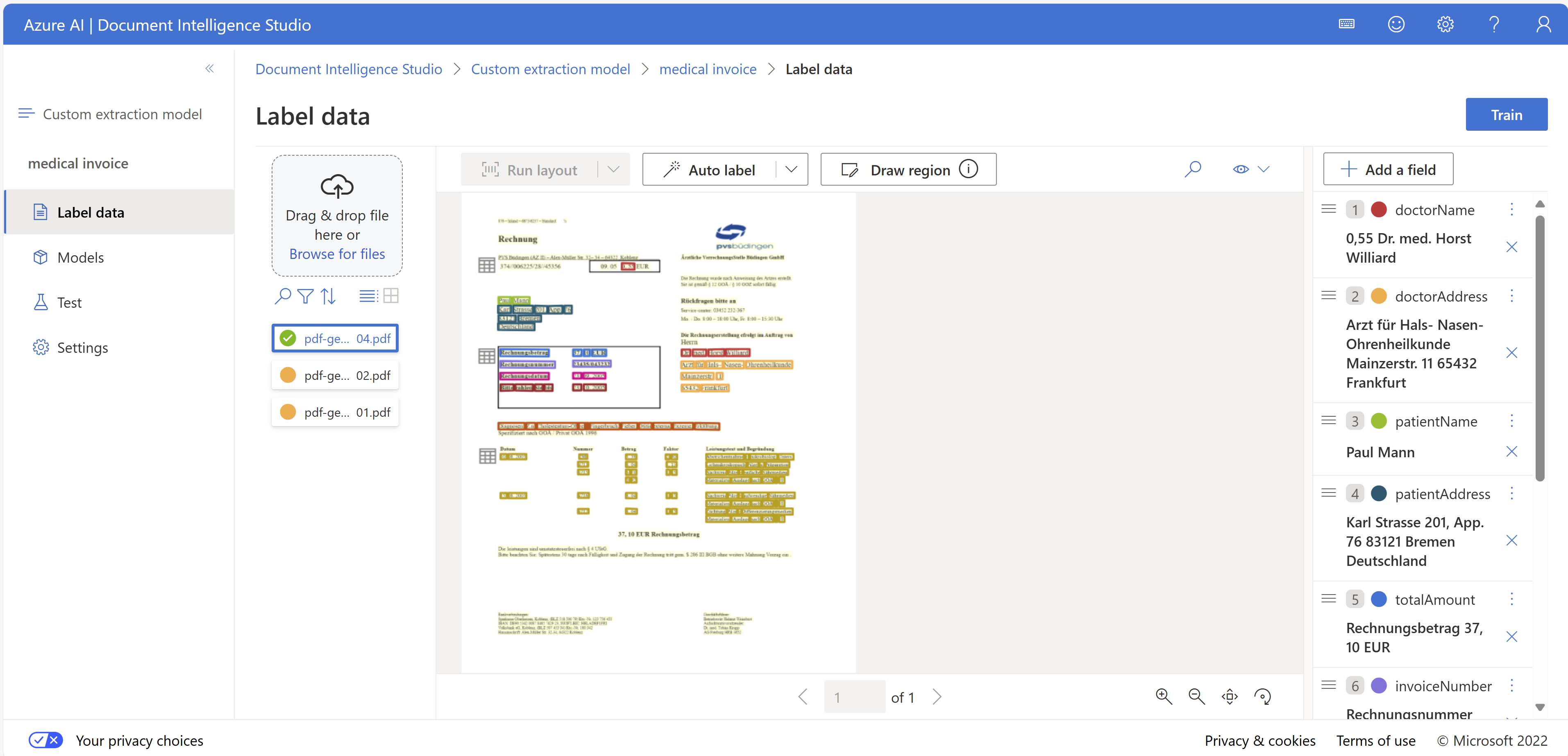
Azure offers several document parsing services. I have used Document Intelligence before, and it has performed very well. There are several standard templates available that work right away. You can also train a custom OCR model.
For the invoices, the standard templates did not work so I trained a custom model. However,
DocIntel is an OCR model that processes layouts. If there is a major change in layouts, it may not perform well. You will need to retrain the model with more examples.
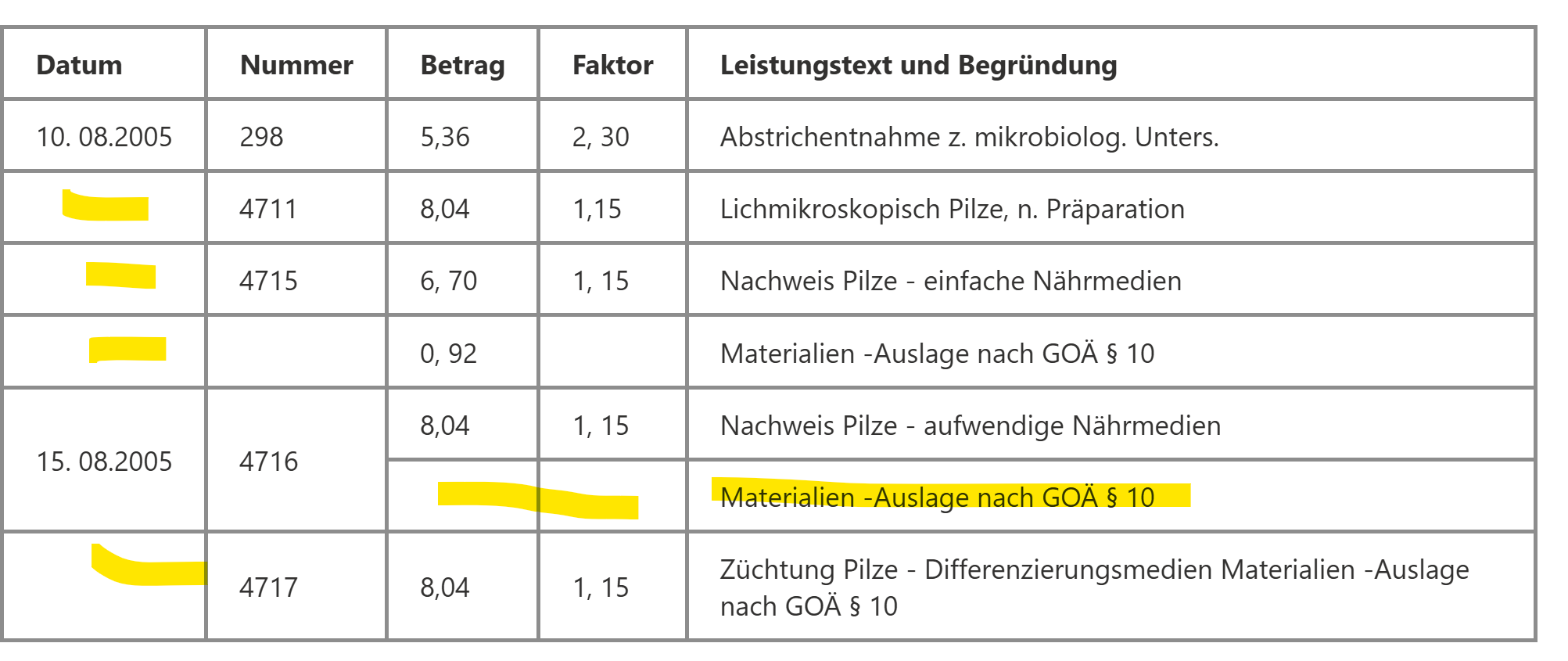
Extracting the items isn't very straightforward because it requires understanding the context. For example, not all items have dates, and the dates need to be "filled down." Item descriptions can continue onto the next line, but DocIntel treats it as a separate line, even though it's supposed to be part of the same item.
If it works, DocIntel is a great tool, and I hope it will be included in the Fabric AI services in the future.
AI Functions
Okay, since the two baselines almost worked, let's see if and how AI Functions can handle this. As I mentioned earlier, this is similar to any other Data Science project, so I:
I split the data for training and testing. Luckily, the data was already divided. The
type01folder contains two folders, each with 20 PDFs. I used PDFs from one folder to adjust the prompt and configurations, and the other for evaluation. I also changed the layout to test for robustness.Extracted the text from the PDFs as markdown. As I have discussed in previous blogs, LLMs love markdwon content. I cleaned up the text and sent it to LLM to extract data using defined JSON schema in
Pydantic.Evaluated based on two simple criteria - schema validation and accuracy.
Schema validation : does the LLM return the data in the defined schema
Accuracy : For the training set, I created the ground truth in Excel and used it for evaluation. Checking the accuracy of the item details is a but tricky so I did that manually and using
deepdiff.
I used
mlflowfor logging the experiment details:extracted json
temperature
input tokens
output token
invalid tokens
schema validation score
accuracy score
prompt
This is an iterative process, so I compared run details and tweaked the prompt/configs until I got the results I wanted.
Like other predictive analytics projects, I started by exploring the solution space. I used a simple prompt and a detailed prompt to understand the model's capabilities for the task. The detailed prompt with a few-shot approach was used to simulate overfitting, and then I scaled back to find the optimal prompt(s). I also did prompt ablation studies.
To keep the blog short, I wont show all the details and like a cooking show skip forward to the final result.
Using .extract :
The easiest way is to use the .extract function for entity extraction based on labels. You can read more about it here.
## Fabric PySpark notebook with runtimr 1.3
%pip install -q pymupdf4llm markitdown[pdf] dateparser deepdiff openai==1.30 > /dev/null 2>&1
%pip install -q --force-reinstall httpx==0.27.0 > /dev/null 2>&1
%pip install -q --force-reinstall https://mmlspark.blob.core.windows.net/pip/1.0.9/synapseml_core-1.0.9-py2.py3-none-any.whl > /dev/null 2>&1
%pip install -q --force-reinstall https://mmlspark.blob.core.windows.net/pip/1.0.10.0-spark3.4-5-a5d50c90-SNAPSHOT/synapseml_internal-1.0.10.0.dev1-py2.py3-none-any.whl > /dev/null 2>&1
Extract pdf text as markdown using pymupdf4llm . You can also use Microsoft’s markitdown library or docling, sparrow or any other library. I have found pymupdf4llm to work well.
Attach a default lakehouse and upload all the PDFs. Change the location path below.
import os
import re
import json
import time
from datetime import datetime
from concurrent.futures import ThreadPoolExecutor
from typing import List, Dict, Optional
import pandas as pd
from tqdm import tqdm
import openai
import pymupdf4llm
from notebookutils import fs
from synapse.ml.aifunc import Conf
from deepdiff import DeepDiff
from pydantic import BaseModel, ValidationError, field_validator
#OPTIONAL if you want to use it instead of pymupdf (check licensinse)
from markitdown import MarkItDown
def create_mkd(path: str):
"""
Convert PDFs to markdown text.
Path should be file path of the attached lakehouse e.g. //lakehouse/default/Files/... if using Fabric
ToDo : Parallelize using ThreadPool in the future, takes ~5min to process all the pdfs currently
"""
try:
# you can use markitdown (OSS by Microsoft) as well
# its very fast but for my use case, didnt work well me with special characters etc
# md = MarkItDown()
# result = md.convert(path).text_content
result = pymupdf4llm.to_markdown(path)
return result, None
except Exception as e:
return None, str(e)
def process_pdfs(path: str):
files = fs.ls(path)
pdf_files = [f for f in files if f.name.endswith(".pdf")] #only pdfs
invoice_data = []
for f in pdf_files:
mkdown_text, error = create_mkd("/lakehouse/default/Files" + f.path.split("Files")[1])
invoice_data.append((f.name, "/lakehouse/default/Files" + f.path.split("Files")[1], mkdown_text, error))
invoice_df = pd.DataFrame(invoice_data, columns=["invoice_name", "invoice_path", "mkdown_text", "mkd_error"])
return invoice_df
def clean_mkdwn(invoice_text):
"""
Clean the markdown to remove titles etc.
"""
processed = re.sub(r'##\s+Herrn\s+###', "## ### ", invoice_text)
processed = re.sub(r'##\s+Herrn\s+([^#])', r"## \1", processed)
processed = re.sub(r'##\s+Frau\s+###', "## ### ", processed)
processed = re.sub(r'##\s+Frau\s+([^#])', r"## \1", processed)
processed = re.sub(r'�', '', processed)
return processed
path = "abfss://Sandeep_Fabric@onelake.dfs.fabric.microsoft.com/aifunc.Lakehouse/Files/dataset-doctor-bills/pdfs/type01/type01b" #1/type01a
invoice_df = process_pdfs(path)
invoice_df['processed_mkdown'] = invoice_df['mkdown_text'].apply(clean_mkdwn)
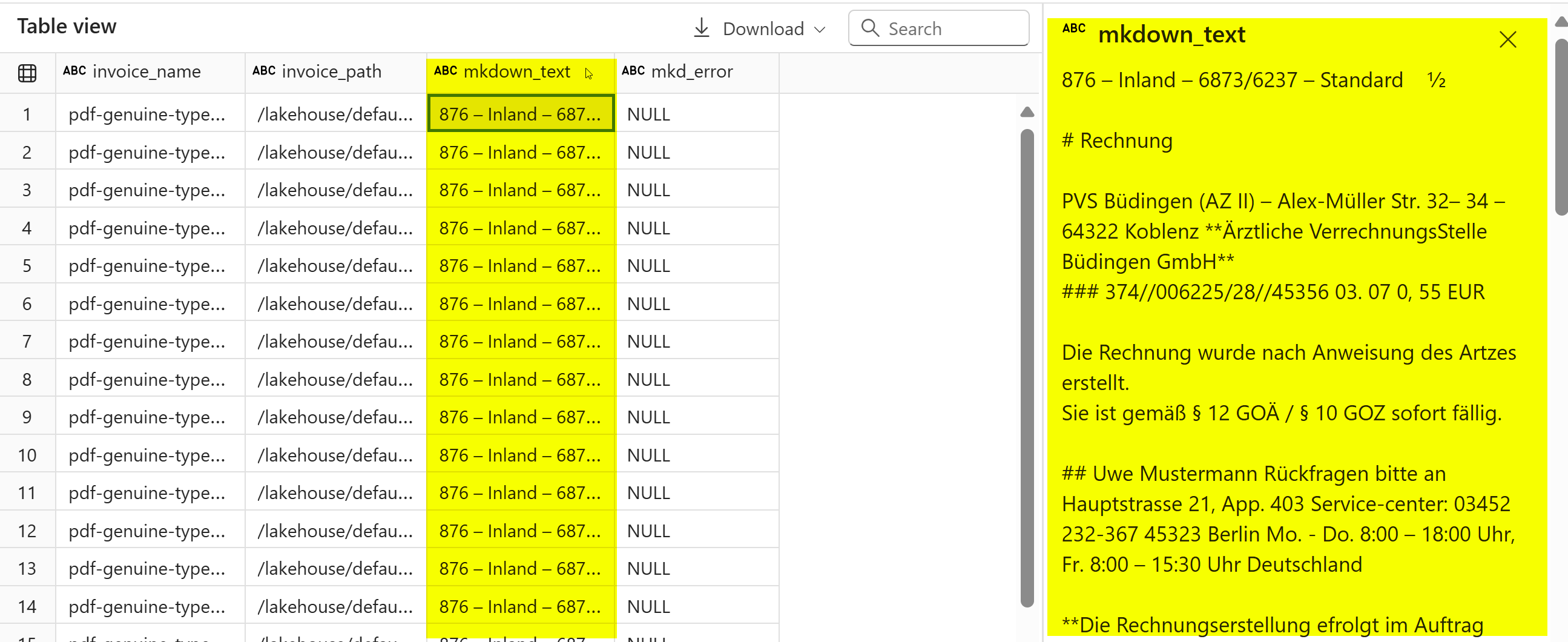
display(invoice_df)
PDFs are extracted as markdown, and cleaned for LLM to work on it.
invoice_df['processed_mkdown'].ai.extract( 'doctor_name',
'doctor_specialization',
'doctor_address',
'doctor_city',
'doctor_postal_code',
'patient_name',
'patient_address',
'patient_city',
'patient_postal_code',
'patient_country',
'invoice_number',
'invoice_date',
'invoice_due_date',
'invoice_total_amount',
'list_of_medical_diagnoses'
)
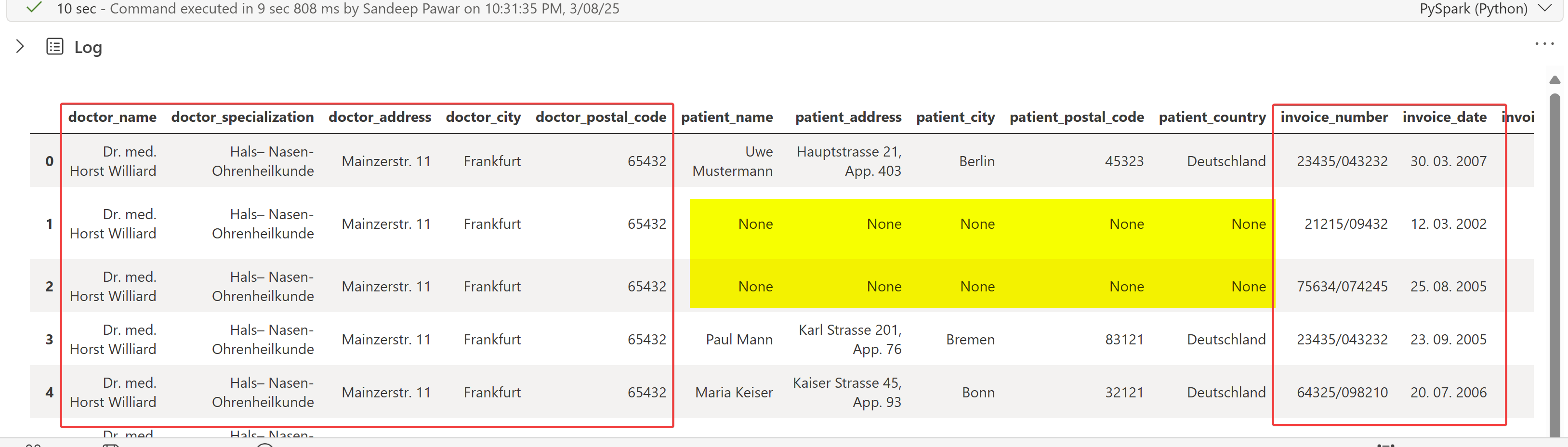
As you can see, with minimal effort, the doctor, invoice details, and list of diagnoses were extracted using just labels, without needing prompts! This is very convenient. However, some patient details were not extracted, and item details cannot be extracted using just labels. If the entities are well-defined, extract() is a great function.
Using .generate_response
If you need to generate a response based on your specific instructions or prompt, you can use the .generate_response function. This function offers a very convenient way to call the AOAI Fabric endpoint. In the following code blocks, I will define the prompt (the final optimized version after testing several options), validate the extracted JSON, clean the text (such as dates, amounts, and removing special characters), and extract item details. Again, I am providing only the final version without the MLflow testing harness and all the prompts I tested.
Prompt
The prompt uses XML tags or instructions for the model. In this case, I am using the gpt-35-turbo model, which I believe doesn’t leverage the tags like the latest models such as gpt-4o and claude 3.5 do. However, it didn't negatively affect the results, so I kept it (I initially experimented using gpt-4o-mini). The prompt structure is similar to what I have discussed in my previous blogs: provide the expected output first, followed by specific yet brief instructions with an example and constraints. In this case, I created a synthetic example to avoid overfitting to a specific example and to cover diverse use cases. Prompt 1 is the basic version with a defined schema, and Prompt 2 is the final optimal prompt. I highly recommend checking out this paper for prompt patterns for data extraction.
prompt1="""
You are a specialized data extraction assistant for German medical invoices. Your task is to extract structured data and convert it to properly formatted JSON shown below
"doctor": {"name", "specialization", "address", "city", "postal_code"},
"patient": {"name", "address", "city", "postal_code", "country"},
"invoice": {"number", "date", "due_date", "total_amount"},
"diagnosis": ["text"],
"items": [{"date", "number", "amount", "factor", "description"}]
}
"""
prompt2="""
<INSTRUCTIONS>
You are a specialized data extraction assistant for German medical invoices.
Your task is to accurately extract structured data and convert it to properly formatted JSON.
Accuracy, consistency, and avoiding hallucinations are crucial for this task.
</INSTRUCTIONS>
<EXPECTED_OUTPUT>
{
"doctor": {"name", "specialization", "address", "city", "postal_code"},
"patient": {"name", "address", "city", "postal_code", "country"},
"invoice": {"number", "date", "due_date", "total_amount"},
"diagnosis": ["text"],
"items": [{"date", "number", "amount", "factor", "description"}]
}
</EXPECTED_OUTPUT>
<RULES>
- Preserve spaces/punctuation EXACTLY
- Precisely extract the doctor's specialization
- Numbers as strings ("5, 00" not 5.00)
- Extract postal code separately from city (e.g., "65432" and "Frankfurt" and not "65432 Frankfurt" )
- For patient's name, look for a person's name other than Doctor's name
- Multi-line item descriptions → \n separation
- Null for missing single values
- Extract only the medical diagnosis from the text. Do not include any billing information, fee schedules (e.g. 'Spezifiziert nach GOÄ / Privat GOÄ 1996'), or other non-diagnostic details.
- Output ONLY valid JSON
</RULES>
<ITEM_PARSING>
Item details contain multiple lines and should be extracted based on following example:
<ITEM_EXAMPLE>
**Datum** **Nummer** **Betrag** **Faktor** **Leistungstext und Begründung**
23. 02. 07 1 10, 72 2, 30 Beratung – auch mittels Fernsprecher
6 13, 41 2, 30 Untersuchung eines Organsystems
2 X 1415 24, 40 2, 30 Binokularmikroskopie
1418 24, 13 2, 30 Endoskopie, Nasen-/Rachenraumm
410 26, 81 2, 30 Sonographie eines Organs B : NNH
04. 03. 07 4722 8, 04 1, 15 Lichtmikroskopische Identifizierung, Pilz
2 X 623 29, 38 1, 80 Thermographie
410 26, 81 2, 30 Sonographie eines Organs Uterus
3 X 420 32, 16 2, 30 Sonographie, je Organ, bis zu drei Blase/bd. Adnexe
4715 6, 70 1, 15 Generic text for demonstration 0, 92 Materialien -Auslage nach GOÄ § 10
</ITEM_EXAMPLE>
<ITEM_JSON>
[
{"date": "23. 02. 07", "number": "1", "amount": "10, 72", "factor": "2, 30", "description": "Beratung – auch mittels Fernsprecher"},
{"date": "23. 02. 07", "number": "6", "amount": "13, 41", "factor": "2, 30", "description": "Untersuchung eines Organsystems"},
{"date": "23. 02. 07", "number": "2 X 1415", "amount": "24, 40", "factor": "2, 30", "description": "Binokularmikroskopie"},
{"date": "23. 02. 07", "number": "1418", "amount": "24, 13", "factor": "2, 30", "description": "Endoskopie, Nasen-/Rachenraumm"},
{"date": "23. 02. 07", "number": "410", "amount": "26, 81", "factor": "2, 30", "description": "Sonographie eines Organs\nB : NNH"},
{"date": "04. 03. 07", "number": "4722", "amount": "8, 04", "factor": "1, 15", "description": "Lichtmikroskopische Identifizierung, Pilz"},
{"date": "04. 03. 07", "number": "2 X 623", "amount": "29, 38", "factor": "1, 80", "description": "Thermographie"},
{"date": "04. 03. 07", "number": "410", "amount": "26, 81", "factor": "2, 30", "description": "Sonographie eines Organs\nUterus"},
{"date": "04. 03. 07", "number": "3 X 420", "amount": "32, 16", "factor": "2, 30", "description": "Sonographie, je Organ, bis zu drei\nBlase/bd. Adnexe"},
{"date": "04. 03. 07", "number": "4715", "amount": "6, 70", "factor": "1, 15", "description": "Generic text for demonstration\n0, 92 Materialien -Auslage nach GOÄ § 10"}
]
</ITEM_JSON>
<ITEM_RULES>
- New Item: Rows with Number, Amount, and Factor columns are new items
- Use row's Date if present, otherwise reuse most recent date
- Description Continuation: Lines missing Date, Number, and Factor are continuations of previous item's description (use \n separator)
- Partial Continuation: Lines with no Date but with Number, Amount, and Factor reuse last date
- Lines with only text are continuations of previous description
</ITEM_RULES>
</ITEM_PARSING>
"""
Generate response: With prompt1, the input tokens = 22868, output tokens = 15231 which is helpful for optimizing the prompt (to keep it within the context window, not hit the rate limit and keep an eye on the cost).
By default the temp is 0, I set the seed=0 for reproducibility.
temp = 0.0
invoice_df['output'] = invoice_df[['processed_mkdown']].ai.generate_response(prompt1, conf=Conf(temperature=temp, seed=0, max_concurrency=25))
num_unevaluated = invoice_df.ai.stats['num_unevaluated'].max()
prompt_tokens = invoice_df.ai.stats['prompt_tokens'].max()
completion_tokens = invoice_df.ai.stats['completion_tokens'].max()
num_unevaluated, prompt_tokens, completion_tokens

Below pydantic is used to define the schema with validation , e.g. which fields are required, which are optional, their data types, validations (e.g. postal code must be 5 digits long). You can use jsonschema as well.
## Pydantic to define the JSON schema and validate
def validate_postal_code(value: str) -> str:
if not value:
return value
if not re.match(r'^\d{5}$', value):
raise ValueError('Postal code must be a string containing exactly 5 digits')
return value
class Doctor(BaseModel):
name: str
specialization: str
address: str
city: str
postal_code: str
@field_validator('postal_code')
@classmethod
def check_postal_code(cls, v: str) -> str:
return validate_postal_code(v)
class Patient(BaseModel):
name: str
address: str
city: str
postal_code: Optional[str] = None
country: Optional[str] = None
@field_validator('postal_code')
@classmethod
def check_postal_code(cls, v: Optional[str]) -> Optional[str]:
if v is None:
return v
return validate_postal_code(v)
class Invoice(BaseModel):
number: Optional[str] = None
date: Optional[str] = None
due_date: Optional[str] = None
total_amount: Optional[str] = None
class Item(BaseModel):
date: str
number: Optional[str] = None
amount: Optional[str] = None
factor: Optional[str] = None
description: str
class InvoiceData(BaseModel):
doctor: Optional[Doctor] = None
patient: Optional[Patient] = None
invoice: Optional[Invoice] = None
diagnosis: List[str] = []
items: List[Item] = []
# additional_info: Optional[AdditionalInfo] = None
def clean_and_parse_json(json_string: str):
mask = re.search(r'\{.*\}', json_string, re.DOTALL)
if mask:
json_string = mask.group(0)
json_string = re.sub(r'[^\S\n]+', ' ', json_string).strip()
json_string = json_string.replace('–', '-')
json_string = re.sub(r'\s*-\s*', '-', json_string)
json_string = json_string.replace('�', '-')
json_string = json_string.replace('°', 'deg')
json_string = json_string.replace('\\"', '"')
try:
return json.loads(json_string)
except json.JSONDecodeError as e:
print(f"Error decoding JSON: {e}")
return None
def validate_invoice_data(data: dict):
try:
valid_data = InvoiceData(**data)
return valid_data, None
except ValidationError as e:
return None, str(e)
def validate_row(row):
parsed = row["cleaned_json_output"]
if parsed is None:
return None, "JSON parsing failed"
valid, error = validate_invoice_data(parsed)
return valid, error
invoice_df["cleaned_json_output"] = invoice_df["output"].apply(clean_and_parse_json)
invoice_df[["validated_data", "validation_error"]] = invoice_df.apply(lambda row: pd.Series(validate_row(row)), axis=1)
invoice_df["is_valid"] = invoice_df["validation_error"].isnull().astype(int)
json_validation_accuracy = (invoice_df["is_valid"].sum())/len(invoice_df["is_valid"])
display(invoice_df)
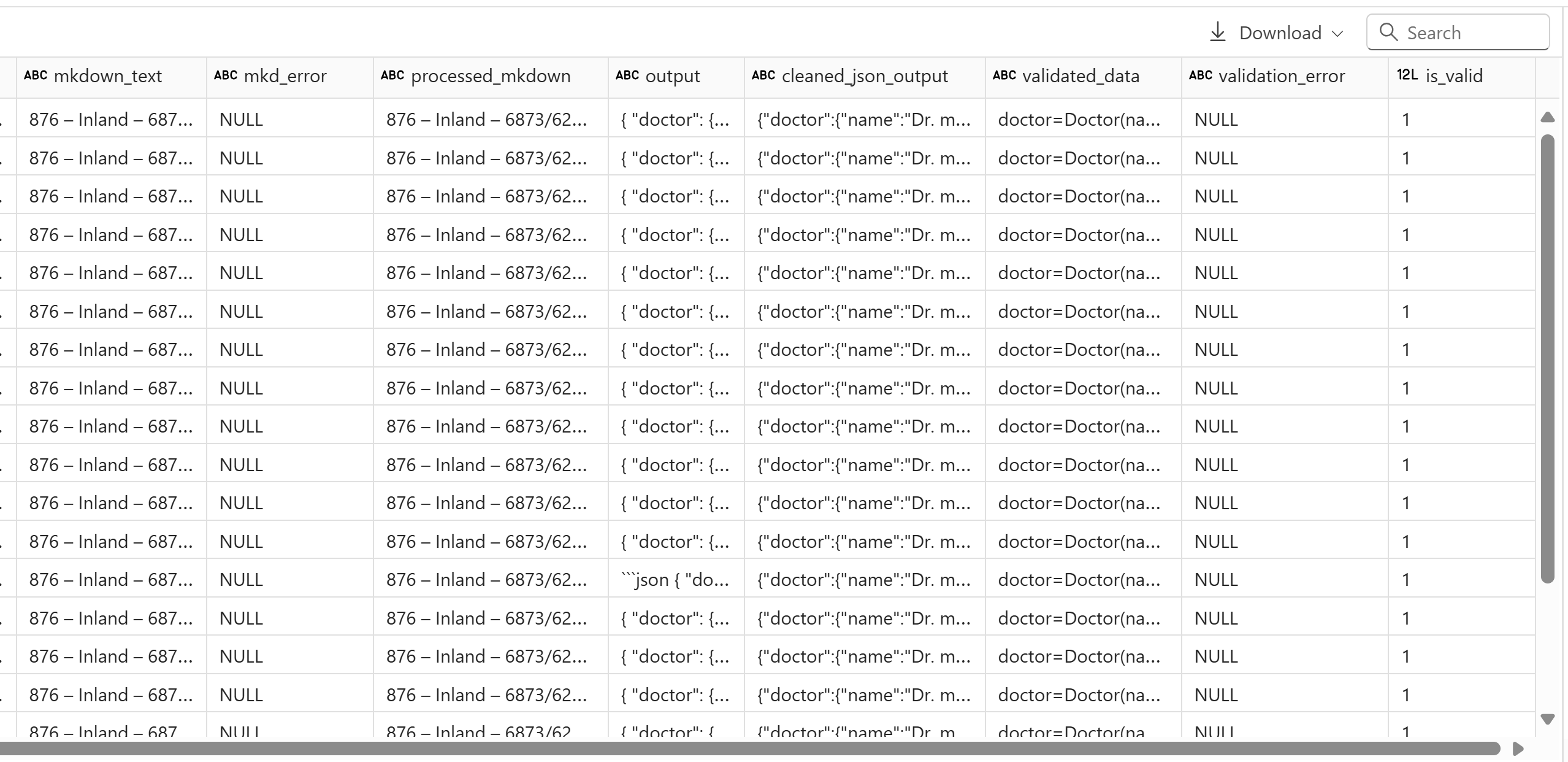
The returned JSON is valid with all the required fields. Next, normalize the JSON to create the dataframe:
def clean_dataframe(invoice_df: pd.DataFrame, json_column:str):
"""
Clean and parse dates and amounts in the DataFrame.
"""
extracted_data = []
for data in invoice_df[json_column]:
extracted_data.append(pd.json_normalize(data))
final = pd.concat(extracted_data)
cleaned_df = final.copy()
date_columns = [col for col in cleaned_df.columns if 'date' in col]
for col in date_columns:
cleaned_df[col] = pd.to_datetime(cleaned_df[col], format='%d. %m. %Y', errors='coerce')
def clean_amount(amount_str):
if pd.isna(amount_str):
return None
cleaned = amount_str.replace('EUR', '').strip()
cleaned = cleaned.replace(',', '.').replace(" ","")
return float(cleaned)
amount_columns = [col for col in cleaned_df.columns if 'amount' in col]
for amt_col in amount_columns:
cleaned_df[amt_col] = cleaned_df[amt_col].apply(clean_amount)
cleaned_df.columns = [x.lower().replace(".","_") for x in cleaned_df.columns]
cleaned_df['invoice_number'] = cleaned_df['invoice_number'].str.replace(" ","")
cleaned_df['clean_diagnosis'] = (cleaned_df['diagnosis'].apply(lambda chars: ", ".join(chars)
.replace("Z.n.", "")
# .replace(" ","")
.replace("[","")
.replace("]","")
.replace("'","")
.replace(" re.","")
.strip())
)
return cleaned_df.reset_index(drop=True)
final_extract = clean_dataframe(invoice_df, "cleaned_json_output")
# translate diagnoses to English
final_extract["english_diagnosis"] = final_extract["clean_diagnosis"].ai.translate("english")
display(final_extract)
Everything is extracted as required - not necessarily accurately.
ai.translate() function to translate the German diagnoses to English which also worked as expected.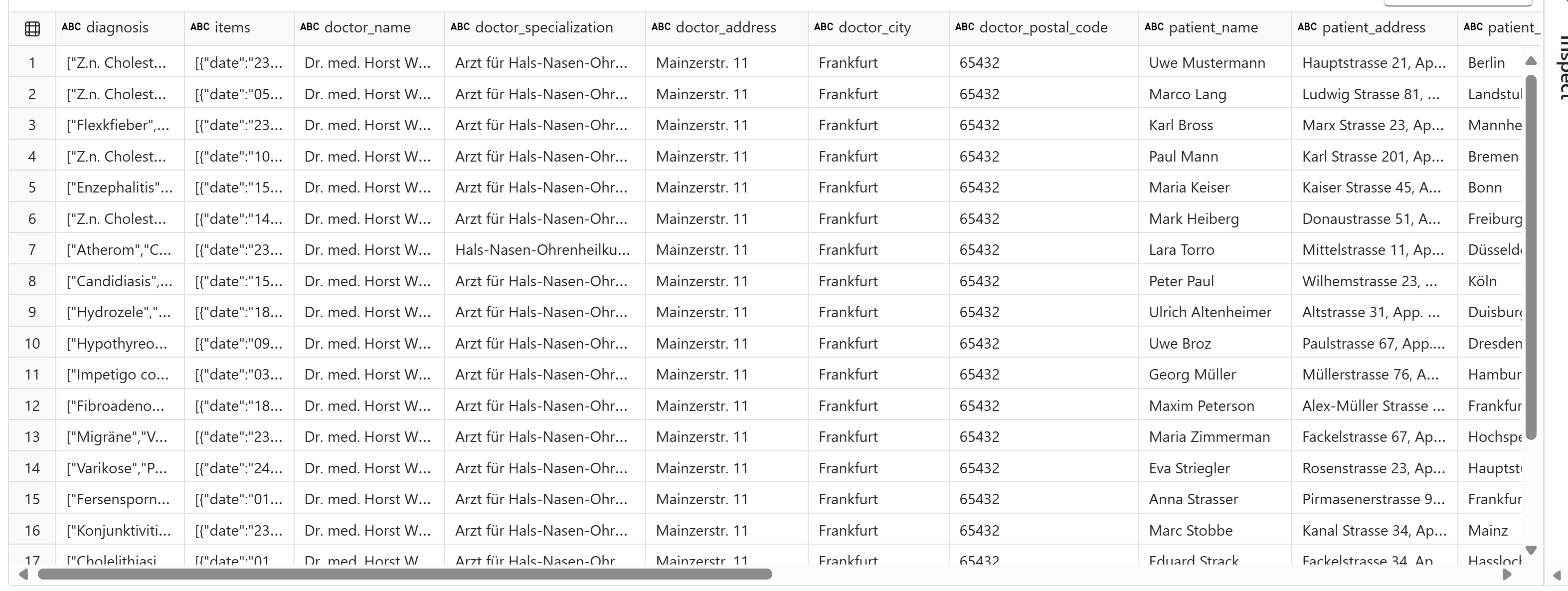
Now, process the items:
import dateparser
from datetime import datetime
import numpy as np
def process_items(final_extract: pd.DataFrame):
"""
Clean, process and create items data
"""
items_data = []
def clean_float(amount_str):
if pd.isna(amount_str):
return None
cleaned = amount_str.replace('EUR', '').strip()
cleaned = cleaned.replace(',', '.').replace(" ", "")
return float(cleaned)
def standardize_date(date_str):
if pd.isna(date_str):
return None
date_str = date_str.replace(" ", "")
parsed_date = dateparser.parse(date_str, languages=['de'])
if parsed_date:
return parsed_date.strftime("%d.%m.%Y")
return date_str
for i, row in final_extract.iterrows():
item_df = pd.json_normalize(row['items'])
item_df['invoice_number'] = row['invoice_number']
item_df['patient_name'] = row['patient_name']
item_df['item_number'] = [row['invoice_number'] + "-" + str(x+1) for x in range(len(item_df))]
items_data.append(item_df)
items_df = pd.concat(items_data, ignore_index=True)
amount_columns = [col for col in items_df.columns if 'amount' in col]
for amt_col in amount_columns:
items_df[amt_col] = items_df[amt_col]
if 'factor' in items_df.columns:
items_df['factor'] = items_df['factor'].str.replace(",", ".").str.replace(" ", "")
items_df['factor'] = pd.to_numeric(items_df['factor'], errors='coerce')
try:
if 'amount' in items_df.columns:
items_df['amount'] = items_df['amount'].str.replace(",", ".").str.replace(" ", "").astype('float')
except Exception as e:
print("-"*50)
print(f"ERROR: {str(e)}")
print("-"*50)
items_df['date'] = items_df['date'].apply(standardize_date)
items_df['description'] = items_df['description'].str.replace("\n", ". ")
items_df['number'] = items_df['number'].str.replace(" . ", ".")
items_df.columns = [x.lower().replace(".", "_") for x in items_df.columns]
return items_df[['item_number', 'invoice_number', 'patient_name','description', 'date', 'number', 'amount', 'factor']].sort_values(['patient_name','item_number']).reset_index(drop=True)
item_df = process_items(final_extract)
display(item_df)
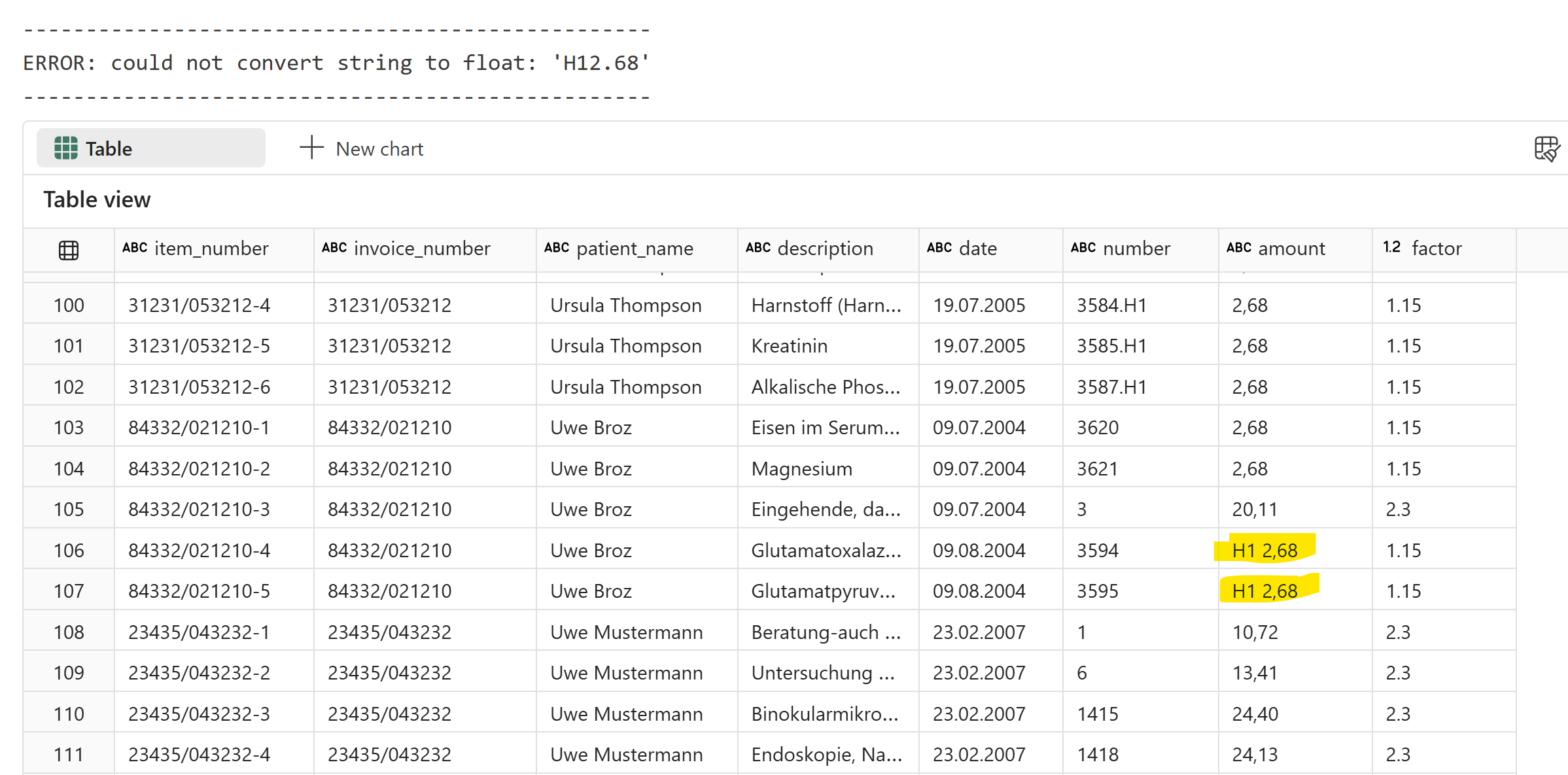
However, I got an error that the value couldn’t be converted because the amount column wasn’t parsed correctly. There are also other issues. The items table is not as straightforward as it looks, here is why:
The number field can be tricky, sometimes the descriptions are split in multiple lines and should be considered as one item.
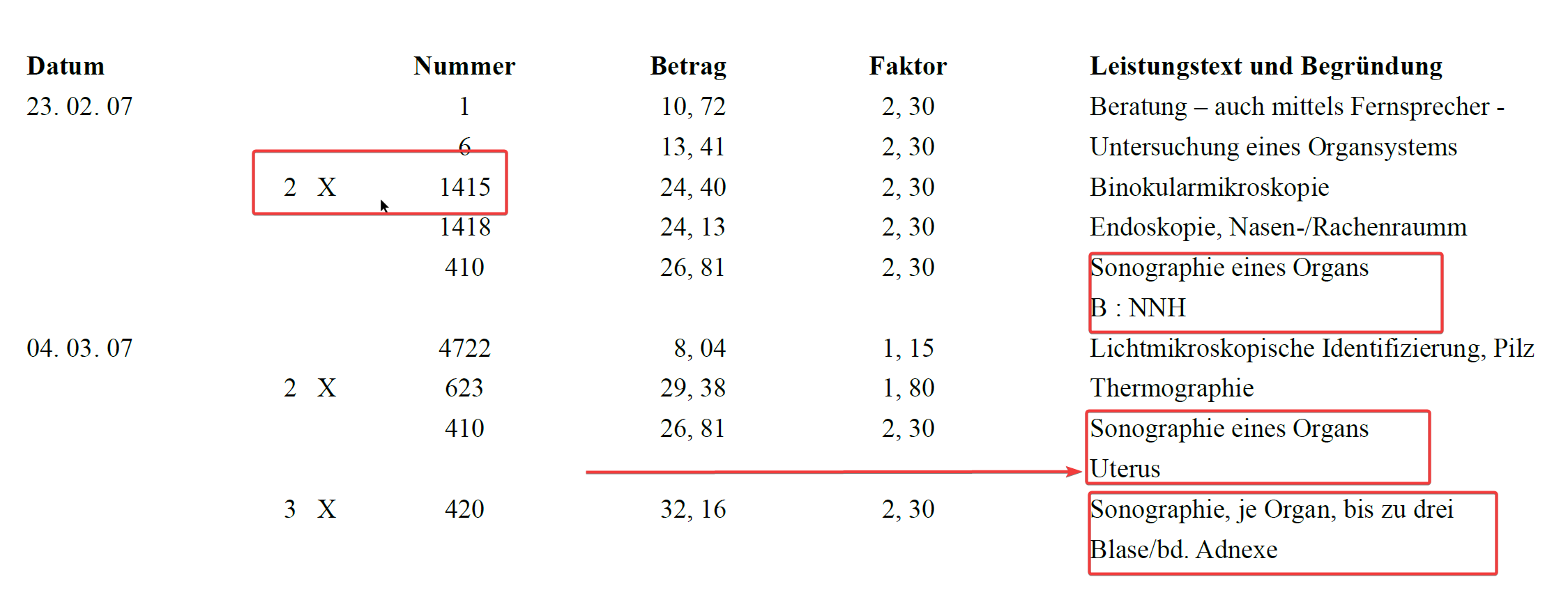
Materialien -Auslage nach GOÄ § 10 can be a line item or it can be a part of an item too depending on the context.
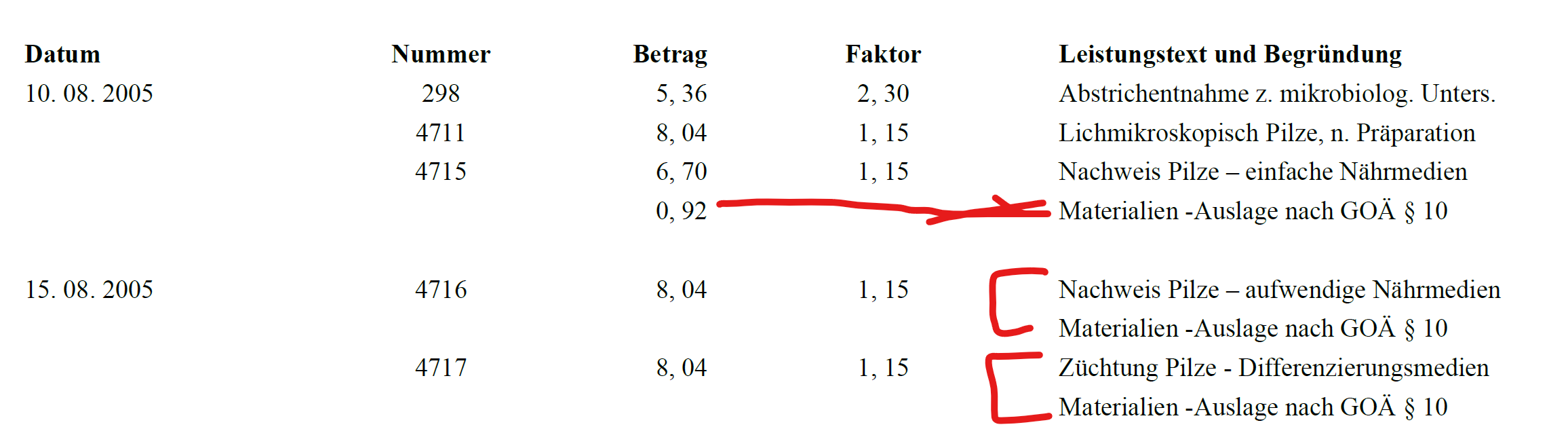
Item descriptions can be multiple lines
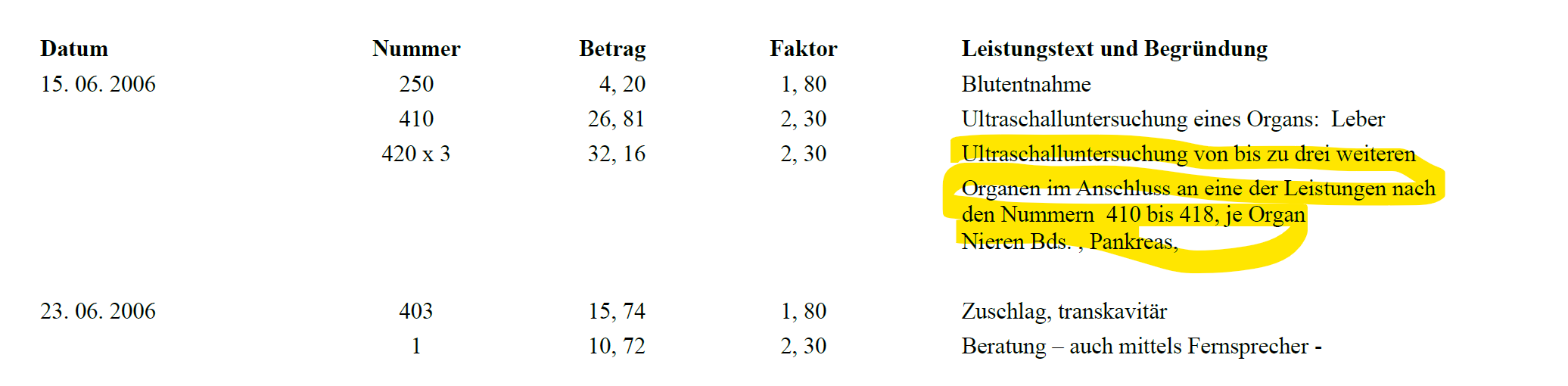
Number and Factor can be blank

Diagnoses have items that are not medical diagnoses.
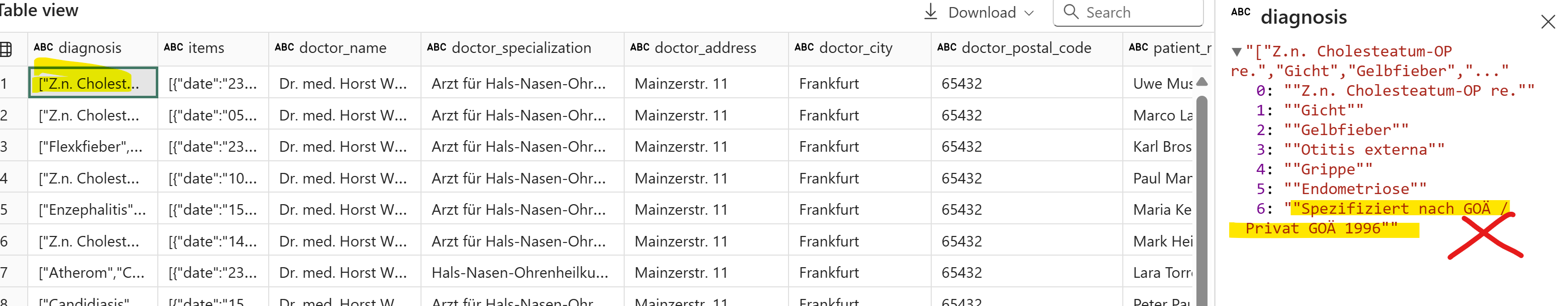
Now, let’s run prompt2 and see what we get:
Prompt 2:
Prompt 2 used twice as many token, 46K input and generated 12K output tokens.
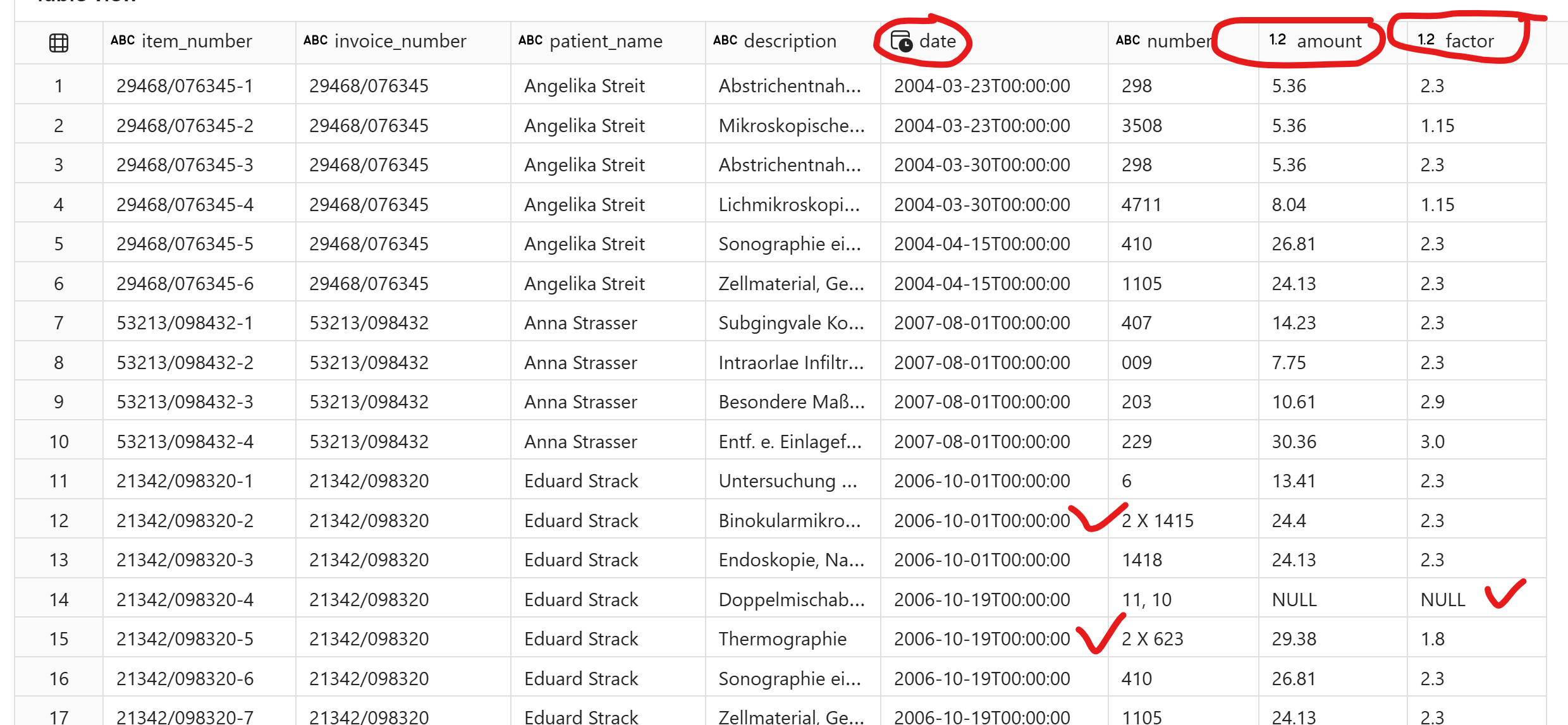
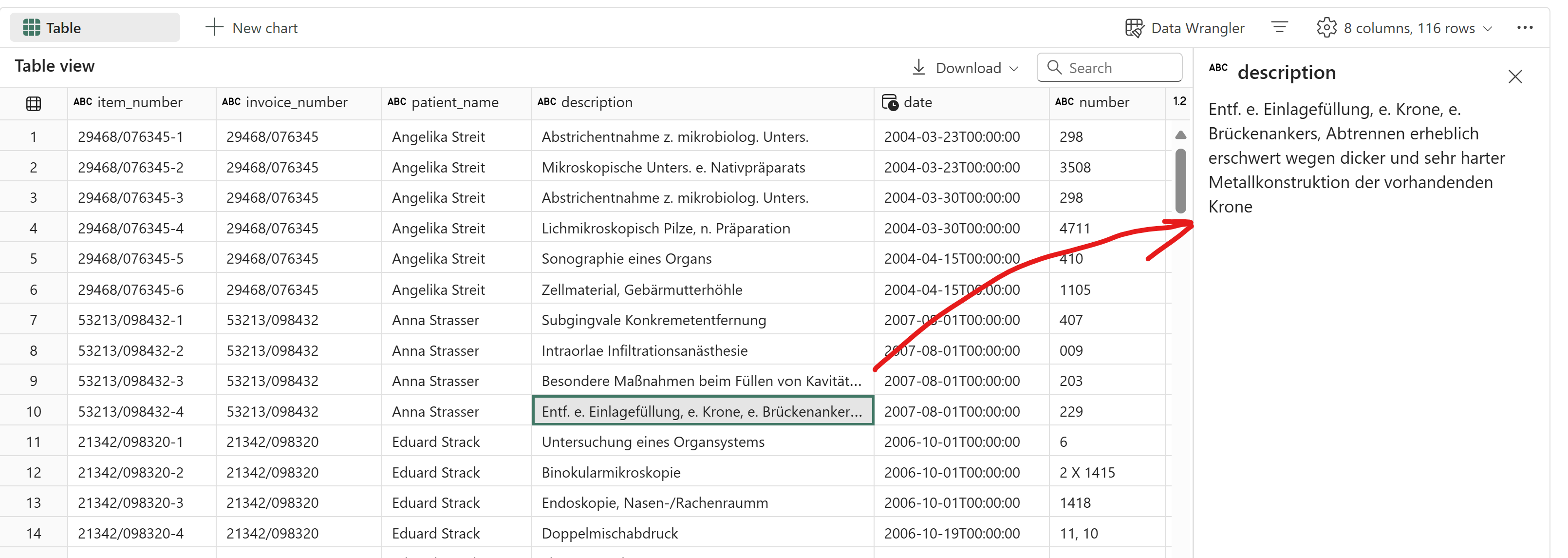
All multi-line descriptions are also captured correctly. Notice that the item dates are also filled down by LLM accurately.
## keep accuracy simple - just compare df w/ ground truth
def calculate_accuracy(source_df, target_df, match_col='is_accurate'):
## compare two dfs for strict validation
result_df = source_df.copy()
if not source_df.columns.equals(target_df.columns):
raise ValueError("Dataframes have different col names")
result_df[match_col] = False
min_rows = min(len(source_df), len(target_df))
for i in range(min_rows):
result_df.loc[i, match_col] = source_df.iloc[i].equals(target_df.iloc[i])
return result_df
## there is a null in one of the date column (correctly) because it's an invalid date, replacing it w/ -1
source_df = final_extract.drop(['diagnosis','items','english_diagnosis'], axis=1).astype('str').replace("NaT","-1")
target_df = pd.read_csv('/lakehouse/default/Files/ground_truth.csv').astype('str').replace("nan","-1")
result = calculate_accuracy(source_df, target_df)
print(f"Overall accuracy: {sum(result.is_accurate)/len(result) *100}%")
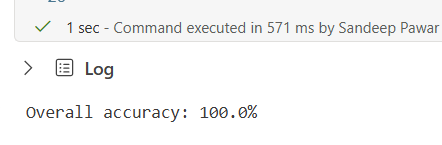
Results are 100% valid and correct on the training set. Download the results from here.
For the sake of completeness, below is a screenshot showing mflow experiment details:
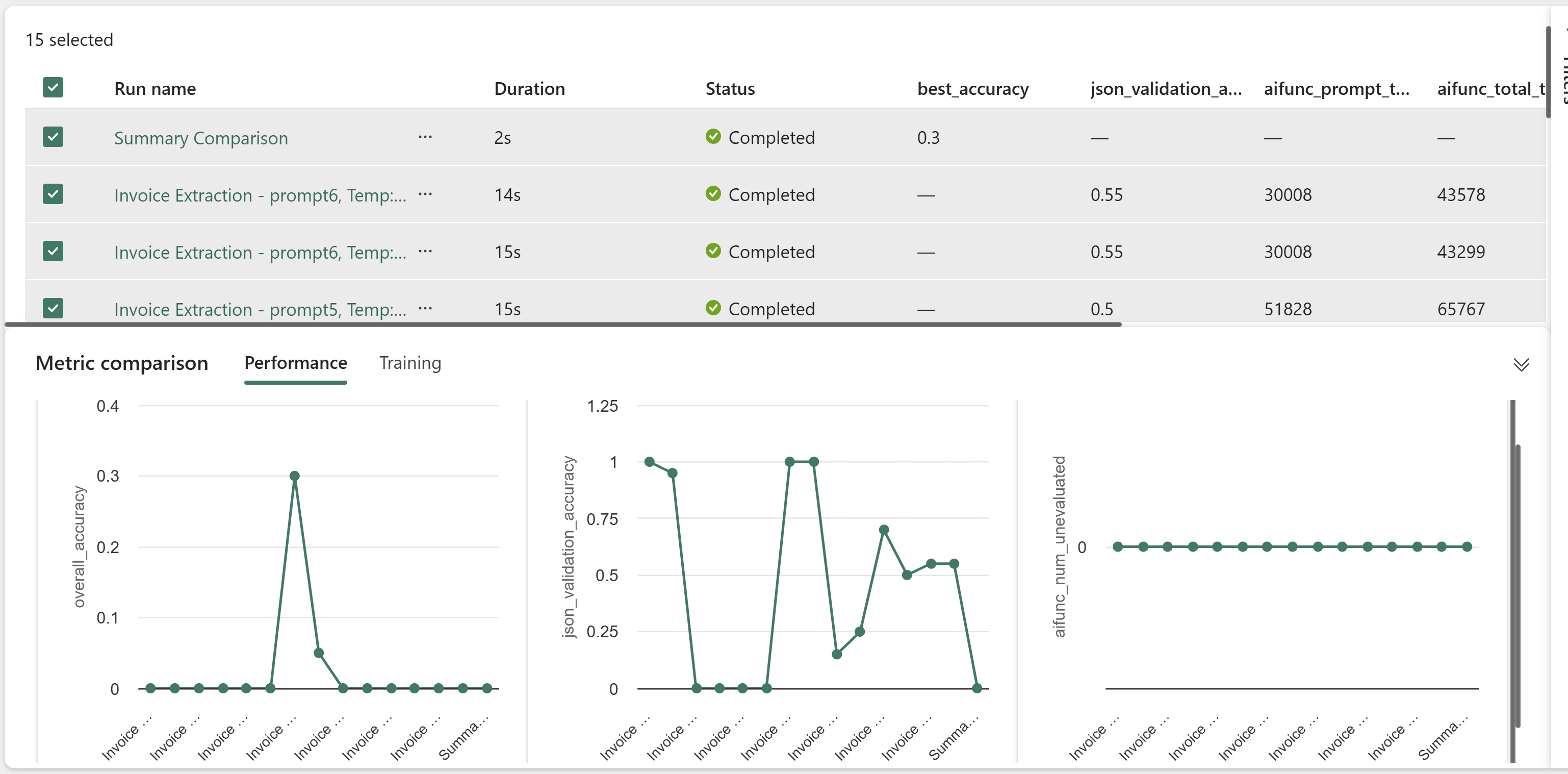
Evaluation
Same layout : All of the above was on the training set. The real test is on the out-of-sample so lets check. These have the same layout as the training set. Everything was valid and accurate.
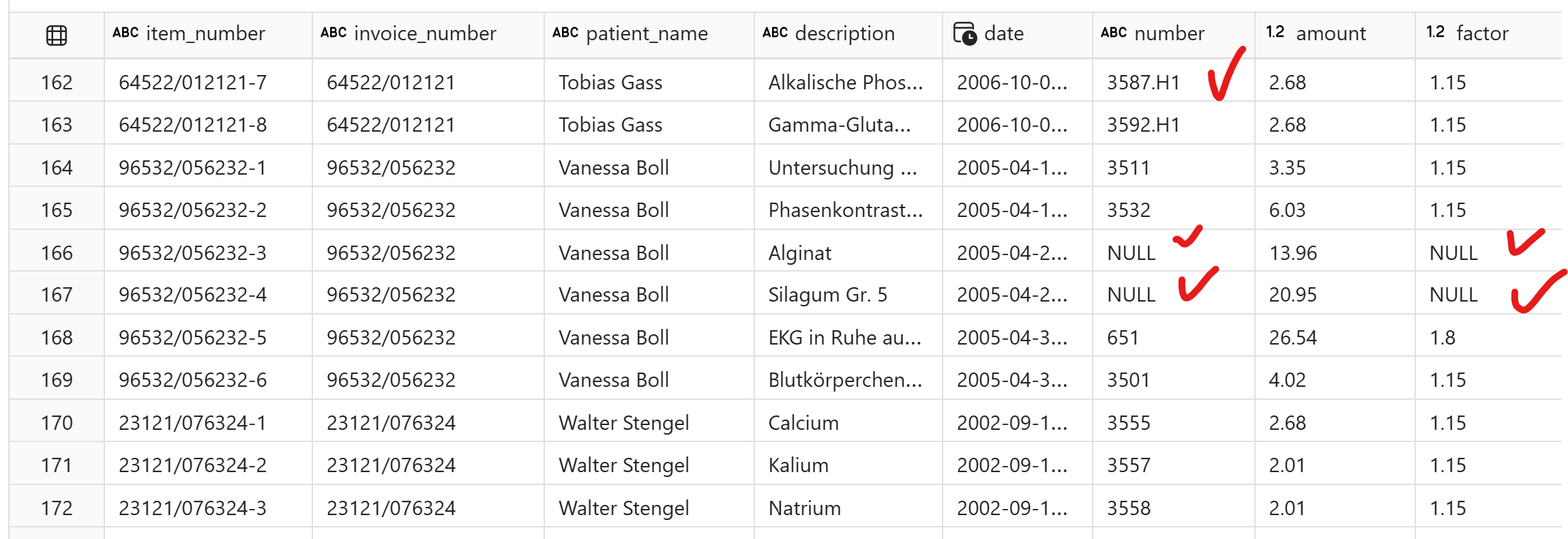
Different layout : For out of distribution test, I re-arranged the text to simulate change in layout to check for robustness:
rearranged = """**Datum** **Nummer** **Betrag** **Faktor** **Leistungstext und Begründung**
13. 07. 2004 3592 . H1 2, 68 1, 15 Gamma - Glutamyltranspeptidase (Gamma Glutamyltransferase, Gamma - GT)
3594 . H1 2, 68 1, 15 Glutamatoxalazetattransaminase (GOT, Aspartataminotransferase, ASAT, AST)
3595 . H1 2, 68 1, 15 Glutamatpyruvattransaminase (GPT, Alaninaminotransferase, ALAT, ALT)
26. 07. 2004 403 4, 53 2, 30 Beseitigung von schrfen Kanten
P3214 150, 00 1, 00 Straumann WN synOcta Abgewinkelt; Sekundärteil; 15 °; Typ A; Höhe 5, 5 m m B; Titan; Art. Nr.: 048.608
P9035 83, 81 3, 70 MainbondSun aufbrennfähig 17005 Bestandteile: 74, 0 % Au; 14, 5 % Ag; 1, 5 % Pt; 5, 5 % Pd; 3, 3 % Zn; 1, 0 % In; 0,1 Ta; 0, 1 % Ir
#### Diagnosen: Zerrung, Cholelithiasis, Chronische Bronchitis, Spezifiziert nach GOÄ / Privat GOÄ 1996
876 Inland 6873 /6237 Standard ½ #
Rechnung PVS Büdingen (AZ II) Alex- Müller Str. 32 34 64322 Koblenz
**Ärztliche VerrechnungsStelle Büdingen GmbH** ### 374 / / 006225 /28 / / 45356 08. 04 0, 55 EUR
Die Rechnung wurde nach Anweisung des Artzes erstellt. Sie ist gemäß § 12 GOÄ / § 10 GOZ sofort fällig.
**Die Rechnungserstellung efrolgt im Auftrag von**
## ### Rechnungsbetrag 246, 38 EUR Dr. med. Horst Williard
Rechnungsnummer 86564 / 053534
Arzt für Hals Nasen - Ohrenheilkunde
Rechnungsdatum 23. 08. 2004 Mainzerstr. 11
Bitte zahlen Sie bis 30. 09. 2004 65432 Frankfurt
## Patrick Brombeerg
Rückfragen bitte an Leipzig Strasse 22, App. 342
Service - center: 03452 232 - 367
42313 Leipzig
Mo. - Do. 8:00 18:00 Uhr, Fr. 8:00 15:30 Uhr Deutschland
### 246, 38 EUR Rechnungsbetrag
Die leistungen sind umstatzsteuerfrei nach § 4 UStG.
Bitte beachten Sie: Spätestens 30 tage nach Fälligkeit und Zugang der Rechnung tritt gem. $ 286 III BGB ohne weitere Mahnung Verzug ein.
Bankverbindungen: Geschäftsführer:
Sparkasse Oberhessen, Koblenz, (BLZ 518 500 79) Kto.-Nr. 123 756 453
Betriebswirt Helmut Weissbrot
IBAN: DE90 5342 0087 8465 7429 23, SWIFT-BIC: HELADEF1FRI
Aufsichtsratsvorsitzender: Volksbank eG, Koblenz, (BLZ 507 453 34) Kto.-Nr. 180 342
Dr. med. Tobias Krupp
Hausanschrift: Alex - Müller Str. 32 - 34, 64322 Koblenz
AG Freiburg HRB 3652 |374 / / 006225 /28 / / 45356|08. 04 0, 55 EUR| |---|---|
"""
invoice_df = pd.DataFrame({"processed_mkdown":[rearranged]})
Everything was valid and extracted correctly:

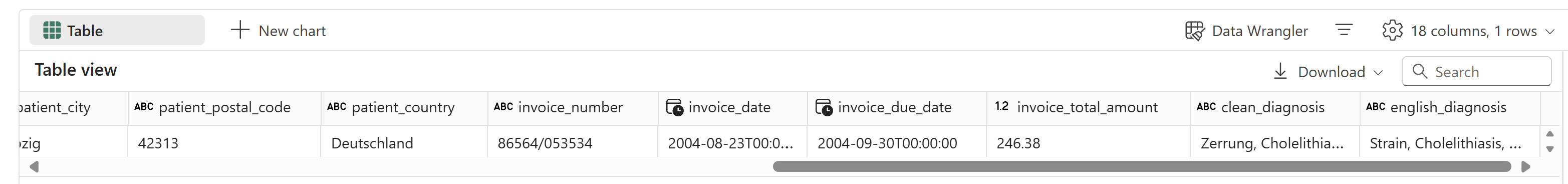
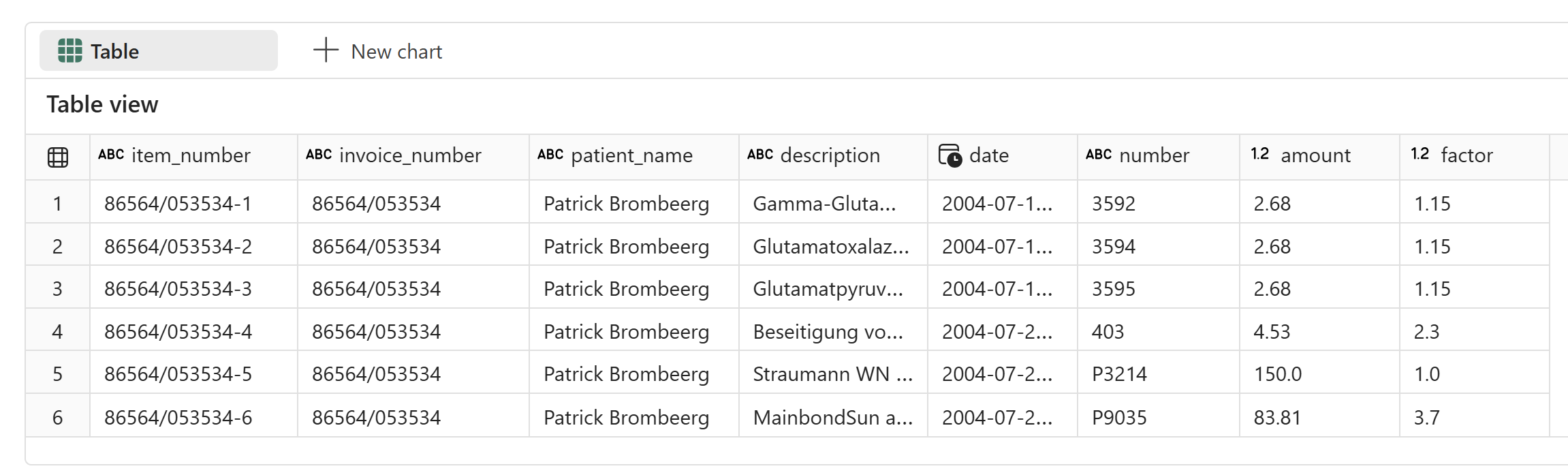
This method should also handle layout changes, which might not be possible with manually labeled OCR-based models. Our AI function is ready for use with production data. You can use .classify or a traditional classification model to identify PDFs with layouts different from those used in training. This is important for maintaining accuracy and detecting drift.
Notes and Observations:
By far the most important thing is understanding the data/patterns and creating the eval harness. Once you have set that up correctly, the rest falls in place as you go through the process.
Notice the
seedparameter. Unlike an ML model where you can seed it for reproducibility, LLMs inherently are stochastic and seed can be used for some deterministic responses on repeated queries but it’s not a guarantee. To check reproducibility, run the queries multiple times with a fixed seed and lower temperature -another reason why creating an eval harness is important to look for even slightest changes. One good proxy for this is tracking the number of output tokens.By default, AI Function uses temperature = 0. You can change it globally per session or by each function using custom configuration. For extraction lower temperature is better to control randomness but for summarization you may want a higher temperature. temp:[ 0,2 ]
I didn’t show field level eval to keep it simple. I used
deepdiffto compare JSON at granular level which was super helpful for identifying variabilities in the responses.I didn’t cover prompt ablation here but my process was to start with a simple prompt and a lengthy prompt and see that works and what doesn’t. Then optimize from there to improve accuracy while keeping the token count down. To identify critical parts of the prompt, I dropped various sections of the prompt and evaluated the responses to identify what’s essential. As you switch to a different models or get new samples, logging these details in the experiment helps in the long run.
As mentioned above, you have always had above capabilities with AOAI Fabric endpoint and Cognitive Services but AI Functions make it super easy to use them.
By default, AI Functions uses
gpt-35-turbo-0125model with 16K context window and Sept 2021 knowledge cutoff. The speed at which LLMs are evolving, that feels like many moons ago. As you saw above, its still a very capable model for this task but I think this a great opportunity for Microsoft to make the recent + faster/cheaper (if not the latest) models available in the Fabric endpoint and differentiate itself given their partnership with OpenAI. They should also make the Phi models available (text as well as multi modal). I do think this will help with Fabric + GenAI adoption especially with convenient features like AI Function to democratize use of GenAI.I absolutely would LOVE to see Azure AI Foundry as a workload in Fabric. This would be a significant catalyst for GenAI discussion and usage in businesses and put Microsoft at the forefront of AI/BI evolution.
If you want to use other Open AI models, you can deploy them to your Azure AI Foundry endpoint and use the custom endpoint in AI Functions, which is great.
If you want to use
gpt-4,gpt-4omodels, you can use Fabric AI services.For complex layouts or many samples, you could use another challenger model to act as a judge and evaluate the responses. This can especially help with automation and quarantining suspect responses.
The consumption rate for this model is 16.81 CU seconds/1000 input tokens and 50.42 CU seconds/1000 output tokens. If my math is right, that’s $0.002 per document for above task ( in RI, PAYG would be 40% more expensive), excluding the compute and storage cost which is negligible. The consumption rate is same as what you would pay in AOAI ($0.5/1M input, $1.5/1M output), no additional discounts in Fabric. This cost would be >1/3rd if/when
gpt-4o-minibecomes available in Fabric endpoint. (Check this reddit discussion for comments by the product team). (My 2c : as I mention above, if you have an RI capacity, you are paying exactly the same rate as paying for your own custom endpoint in AOAI. So if you have use cases for AI Functions, you are NOT paying anything extra. In fact, I would have loved to see it discounted in Fabric. If you are concerned about capacity being chewed, you could use your endpoint to monitor/control costs and control rate limits etc.. On the other hand, if you are using a PAYG, you are better off by not using the default Fabric endpoint because its 40% more expensive, in that case definitely use the custom endpoint. I love that the product team made this option available!)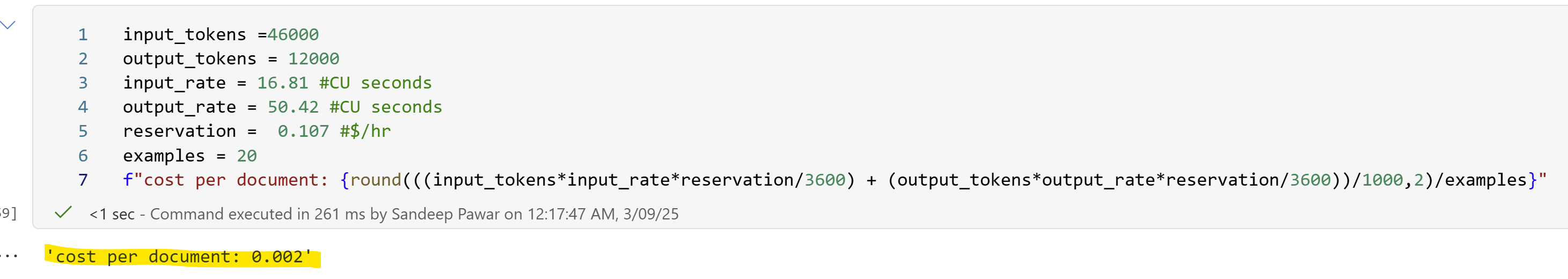
Rate limit for this model is 1000 RPM which is ~160K token/min which should be good for ~50 samples/min. If you have hundreds or thousands of samples, this will be a challenge (correct if I am wrong, please). Again, this limitation may be removed in the future.
Unless you define the custom endpoint, I believe AI Functions only work in PySpark notebook for now. I would expect that to be a short-term gap. Also notice that you have to pip install it. Once the new Fabric runtime is rolled out, it will likely be part of it.
Similar to PDF, you can also process Excel, audio, YT transcripts, emails, Word documents etc. This was rather a simple example tbh. Check out
markitdownlibrary by Microsoft (I have included a snippet in the code above).Fabric also has Osmos AI Data Wrangler workload (3rd party) which uses agentic AI to process more complex documents. I haven’t tried it yet but it’s on my list.
If you need to handle more complex documents, forms etc, using an API I recommend trying NuExtract and Gemini Flash 2.0. Both so far have worked exceedingly well for me.
Above method doesn’t work if your document has hand-written text, radio buttons, check boxes etc. For that either try a vision based model or
Marker,docling,sparrow,llamaextract.Similar to other AI services in Fabric, you need paid F64+ capacity to use AI Functions. For <F64, you can use a AOAI custom endpoint and create custom config.
I am not 100% sure ifFabric Copilot Capacitycan be used for <F64.FCC cannot be used for AI Functions, you have to have F64+. If you don’t have complex use cases, you could also save an OSS LLM model to a lakehouse and use it for inferencing.e.g. I used a quantized version ofphi-4-minilocally in Fabric and it worked well (more on this later in a future blog). However, since Fabric does not have GPUs, I am using the ONNX version on CPU and it’s slow.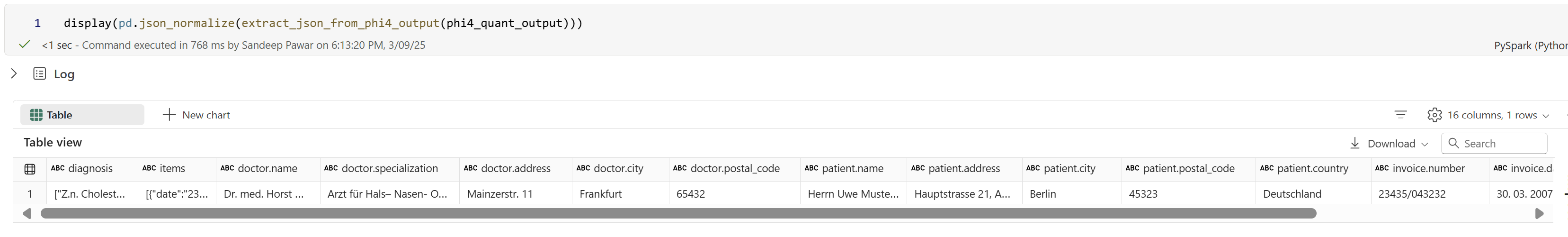
If would like to learn more about AI features in Fabric, please attend my session at upcoming FabCon:
References:
Use Azure AI services in Fabric - Microsoft Fabric | Microsoft Learn
Transform and enrich data seamlessly with AI functions - Microsoft Fabric | Microsoft Learn
Best practices for prompt engineering with the OpenAI API | OpenAI Help Center
Work with the GPT-35-Turbo and GPT-4 models - Azure OpenAI Service | Microsoft Learn
microsoft/markitdown: Python tool for converting files and office documents to Markdown.
Prompt_Patterns_for_Structured_Data_Extraction_from_Unstructured_Text.pdf
Subscribe to my newsletter
Read articles from Sandeep Pawar directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by

Sandeep Pawar
Sandeep Pawar
Principal Program Manager, Microsoft Fabric CAT helping users and organizations build scalable, insightful, secure solutions. Blogs, opinions are my own and do not represent my employer.