Visualizing and Annotating Protein Structures in 3D with JavaScript: 3DMol
 Biohacker0
Biohacker0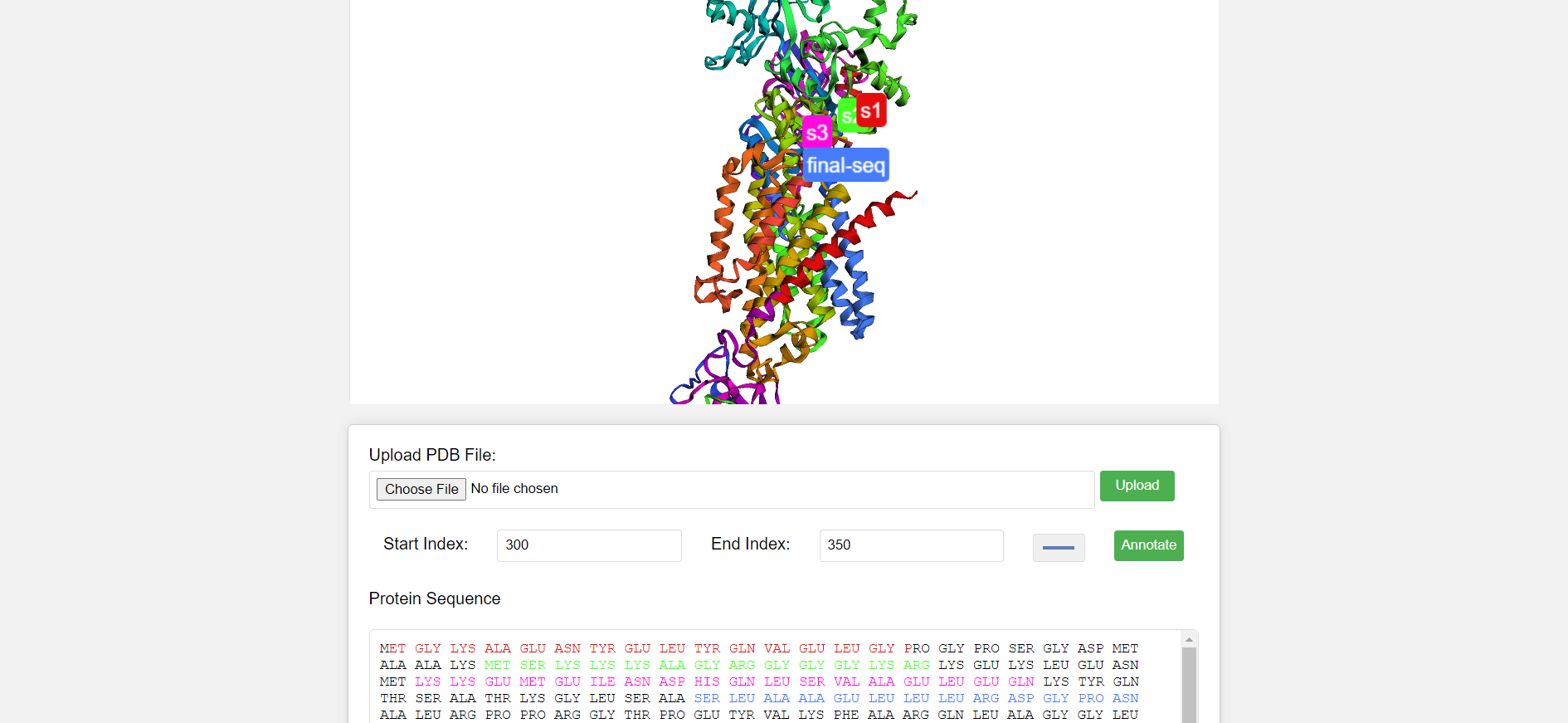
In this project, I've developed a web application for annotating protein sequences based on Protein Data Bank (PDB) files. The application allows users to upload PDB files, visualize protein structures, select specific segments within the structure, and annotate them with customized colors. Additionally, it displays the protein sequence extracted from the uploaded PDB file and highlights the annotated segments within the sequence.
Objectives:
1. Loading and Displaying PDB Structures:
Reads PDB files, either a default one or user-uploaded.
Renders the protein structure in a 3D viewer using the $3Dmol.js library.
Extracts and displays the amino acid sequence.
2. Interactive Annotation:
Allows users to select regions of the protein structure.
Highlights selected regions in chosen colors within the 3D visualization.
Adds labels with custom names to the highlighted regions.
Highlights corresponding amino acids in the sequence display.
# Theoretical Foundations:
PDB (Protein Data Bank) Files: Standard format for storing 3D structures of proteins and other biological molecules.
$3Dmol.js: A JavaScript library for interactive 3D molecular visualization. - Amino Acid Sequence: The linear order of amino acids that make up a protein, determining its structure and function.
# Core Logic and Function Breakdown:
1. Event Listeners and File Handling:
DOMContentLoaded: Ensures code execution after the web page's content is loaded.pdbFileInput.addEventListener: Handles user-uploaded PDB files.
2. Loading PDB Data:
loadDefaultPDB: Loads a default PDB file if none is uploaded.loadPDBData:Creates a 3D viewer using $3Dmol.js.
Adds the PDB data to the viewer.
Sets initial visualization styles (e.g., color spectrum).
Zooms to fit the protein structure.
Extracts and displays the amino acid sequence.
3. Handling Annotations:
annotateButton.addEventListener: Triggers annotation upon button click.Retrieves user input for start and end indexes of the region to highlight.
Validates input to ensure valid numbers.
Applies the selected color to the specified region in the 3D viewer.
Adds a label with the user-provided name.
Highlights corresponding amino acids in the sequence display.
4. Highlighting Text:
highlightText: Synchronizes highlighted regions in the 3D viewer with the amino acid sequence display.
5. Extracting Sequence:
extractSequence: Extracts the amino acid sequence from the PDB data.
Core Logic /Algorithmic approach:
Function Breakdown:
loadDefaultPDB():
Loads a pre-defined PDB file path from a variable (
defaultPDBFile).Uses jQuery.ajax to fetch the file data asynchronously.
Calls
loadPDBData()with the fetched data upon successful retrieval.Logs an error message to the console if the request fails.
function loadDefaultPDB() { // Load default PDB file if no file uploaded jQuery.ajax(defaultPDBFile, { success: function (data) { loadPDBData(data); }, error: function (hdr, status, err) { console.error("Failed to load default PDB file: " + err); }, }); }
loadPDBData(pdbData):
Clears the existing viewer if one exists.
Creates a new 3D viewer using
$3Dmol.createViewer()and configures its background color.Adds the provided
pdbDatato the viewer usingviewer.addModel().Sets the initial visualization style using
viewer.setStyle(), applying a "spectrum" color scheme to the cartoon representation.Zooms to fit the protein structure using
viewer.zoomTo().Renders the updated viewer using
viewer.render().Calls
extractSequence(pdbData)to retrieve the amino acid sequence.Displays the extracted sequence in the
sequenceDisplayelement.
function loadPDBData(pdbData) {
if (viewer) {
viewer.clear();
}
let config = { backgroundColor: "white" };
viewer = $3Dmol.createViewer(element, config);
viewer.addModel(pdbData, "pdb");
viewer.setStyle({}, { cartoon: { color: "spectrum" } });
viewer.zoomTo();
viewer.render();
let sequence = extractSequence(pdbData);
sequenceDisplay.innerText = sequence;
}
annotateButton.addEventListener('click', ...):
This event listener is triggered when the "annotate" button is clicked.
Parses the user-provided start and end indexes from the input fields.
Validates the input to ensure they are valid numbers.
Applies the selected color to the specified region using
viewer.setStyle().Creates a highlight object with start, end, and color information.
Pushes the highlight object to the
highlightedSegmentsarray.Calculates the average coordinates of the highlighted region for label placement.
Prompts the user for a label name using
prompt().If a label name is provided, adds it to the viewer using
viewer.addLabel(), positioning it at the calculated coordinates and styling it with the selected color.Renders the updated viewer using
viewer.render().Calls
highlightText()to synchronize highlights in the sequence display.
annotateButton.addEventListener("click", function () {
let selectionStart = parseInt(startInput.value.trim());
let selectionEnd = parseInt(endInput.value.trim());
let selectedColor = colorPicker.value;
if (isNaN(selectionStart) || isNaN(selectionEnd)) {
alert("Please enter valid start and end indexes.");
return;
}
viewer.setStyle(
{ resi: selectionStart + "-" + selectionEnd },
{ cartoon: { color: selectedColor } }
);
highlightedSegments.push({
start: selectionStart,
end: selectionEnd,
color: selectedColor,
});
let x = 0,
y = 0,
z = 0;
for (let i = selectionStart; i <= selectionEnd; i++) {
let atom = viewer.getModel().selectedAtoms({ serial: i })[0];
x += atom.x;
y += atom.y;
z += atom.z;
}
x /= selectionEnd - selectionStart + 1;
y /= selectionEnd - selectionStart + 1;
z /= selectionEnd - selectionStart + 1;
let sequenceName = prompt("Enter sequence name:");
if (sequenceName) {
viewer.addLabel(sequenceName, {
position: { x: x, y: y, z: z },
fontSize: 20,
backgroundColor: selectedColor,
});
viewer.render();
}
highlightText();
});
highlightText():
Iterates through each character in the sequence displayed in the
sequenceDisplayelement.Checks if the current character's index falls within any of the highlighted regions stored in
highlightedSegments.If a match is found, wraps the character in a
<span>element with a class of "highlighted" and sets the inline style to match the corresponding highlight color.Otherwise, appends the character directly to the highlighted sequence string.
Updates the inner HTML of the
sequenceDisplayelement with the constructed highlighted sequence.function highlightText() { let sequenceText = sequenceDisplay.innerText; let highlightedSequence = ""; let lastIndex = 0; for (let i = 0; i < sequenceText.length; i++) { let char = sequenceText.charAt(i); let color = null; for (let segment of highlightedSegments) { if (i >= segment.start && i <= segment.end) { color = segment.color; break; } } if (color) { if (lastIndex !== i) { highlightedSequence += sequenceText.substring(lastIndex, i); } highlightedSequence += `<span class="highlighted" style="color: ${color};">${char}</span>`; lastIndex = i + 1; } } if (lastIndex < sequenceText.length) { highlightedSequence += sequenceText.substring(lastIndex); } sequenceDisplay.innerHTML = highlightedSequence; }
extractSequence(pdbData):
Splits the
pdbDatastring into lines usingsplit('\n').Initializes an empty string to store the extracted sequence.
Iterates through each line:
Checks if the line starts with "SEQRES", indicating a sequence definition.
If a sequence line is found, splits it into tokens using
split(/\s+/).Extracts amino acid residues from tokens starting from index 4 and appends them to the sequence string with spaces.
Trims leading and trailing spaces from the extracted sequence and returns it
function extractSequence(pdbData) { let lines = pdbData.split("\n"); let sequence = ""; for (let line of lines) { if (line.startsWith("SEQRES")) { let tokens = line.trim().split(/\s+/); for (let i = 4; i < tokens.length; i++) { sequence += tokens[i] + " "; } } } return sequence.trim(); }
Subscribe to my newsletter
Read articles from Biohacker0 directly inside your inbox. Subscribe to the newsletter, and don't miss out.
Written by

Biohacker0
Biohacker0
I am a software engineer and a bioinformatics researcher. I find joy in learning how things work and diving into rabbit holes. JavaScript + python + pdf's and some good music is all I need to get things done. Apart from Bio and software , I am deeply into applied physics. Waves, RNA, Viruses, drug design , Lithography are something I will get deep into in next 2 years. I will hack biology one day